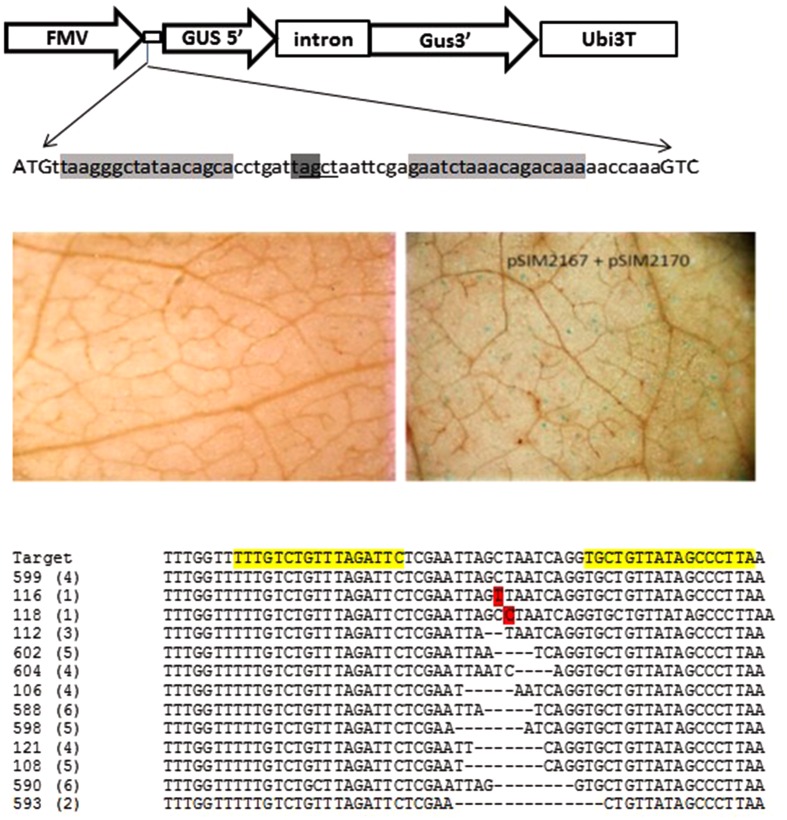
FIGURE 3.
Verification of designed TALENs in N. benthamiana infiltration. (Top panel) Target vector pSIM2167 of TALENs for N. benthamiana infiltration. A 60 bp DNA sequence from StUbi7 5′ intron was inserted between the first and second amino acid codons of the GUS gene. There is an in frame stop codon (dark gray highlighted) in the spacer region between the two recognition sites (light gray highlighted) which will abolish the GUS protein translation. TALEN-mediated DNA cleavage and subsequent NHEJ based repairing will destroy the stop codon and translation of the GUS will be in frame and restore GUS activity. An AluI restriction enzyme site (underlined) was also included in the spacer to facilitate detection of TALEN-mediated indels. (Middle and bottom panels): The functionality of designed TALEN in transient assay. Target construct was Agro-infiltrated into N. benthamiana leaves alone or together with the TALEN construct. Forty-eight hours after infiltration, leaf tissue at the infiltrated site was collected and stained with GUS staining solution and de-stained with ethanol for microscopic examination. DNA was isolated from co-infiltrated tissue, digested with AluI, PCR amplified, cloned and sequenced. (Middle left panel): Target construct pSIM2167 alone. (Middle right panel): Target construct pSIM2167 co-infiltrated with TALEN construct pSIM2170. (Bottom panel): Sequences of target region with various modifications. The number in the parenthesis represents how many times the modification occurred in the 50 clones sequenced. The numbers on the left are identifiers of the respective clones.