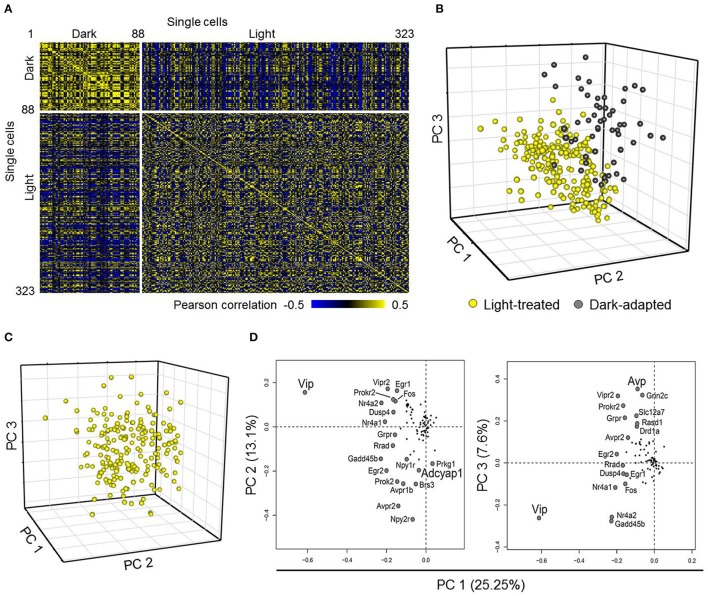
Figure 2.
Comparing neurons from dark-adapted and light-treated mice. (A) Pearson correlation of 332 single neurons collected across dark-adapted (88 neurons) and light-treated (235 neurons) mice. (B) Scores plot obtained from a Principal Component Analysis (PCA) on the single neurons (spheres) sampled shows that neurons responding to the light pulse (yellow) form a distinct cluster from that of the dark-adapted neurons (dark-gray) along the first three principal components. The distinct clusters indicate distinct transcriptional states between these two treatment groups. (C) Scores plot obtained from PCA of only neurons from light pulsed animals shows a large amount of variability as indicated by the large spread of neurons. (D) Loading values obtained from the PCA analysis of neurons taken from light-perturbed animals are plotted for PC1 and PC2 and PC1 and PC3. The labeled genes contribute more to the observed variability in the data, with the signaling neuropeptide genes Vip and Avp having the largest contributions to neuronal variability along PC1 and PC2. Adcyap1 and other labeled genes contribute to variability along PC3.