Figure 2.
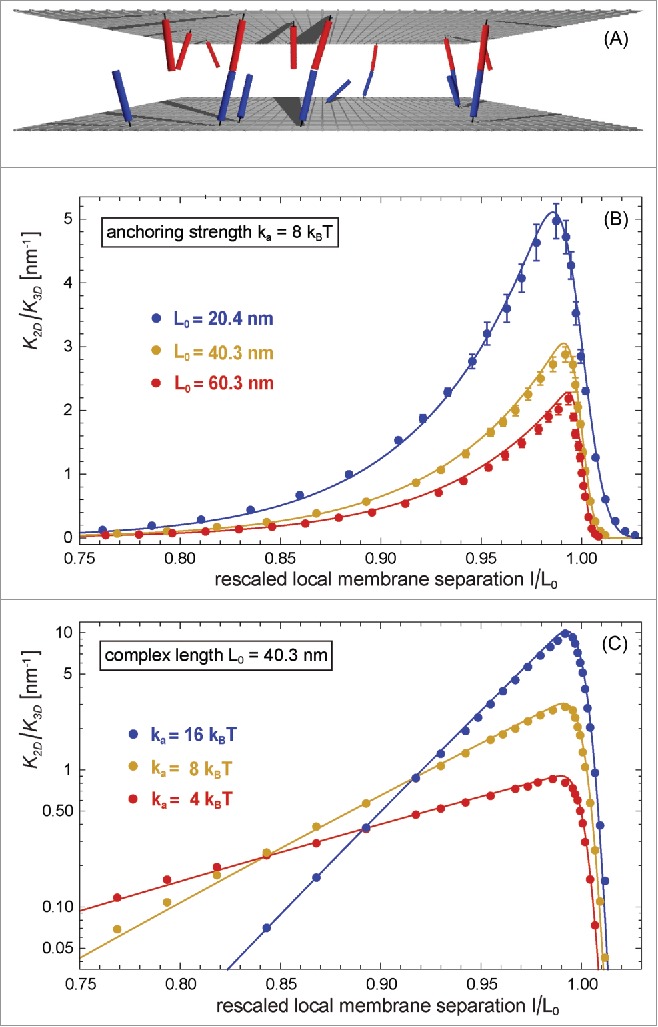
(A) Snapshot from a MC simulation with parallel and planar membranes. (B) and (C) Ratio of the binding constants of membrane-anchored and soluble receptors and ligands versus local membrane separation l for different anchoring strengths and complex lengths of the receptors and ligands of our elastic-membrane model of biomembrane adhesion. The data points represent MC data, and the lines theoretical results based on Eqs. (7) and (8). The binding constant of soluble variants of the receptors and ligand is determined by the binding potential of the receptors and ligands and does not depend on the complex length .
