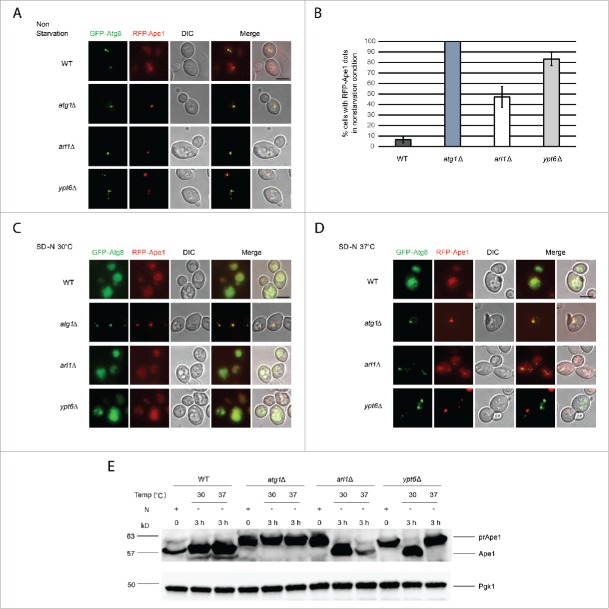
Figure 7.
Arl1 and Ypt6 are required in the Cvt pathway. mRFP-Ape1 was integrated into the yeast genome using the pRS305-mRFP-Ape1 plasmid. Cells were grown and treated under starvation conditions as described. (A) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains in nonstarvation conditions. (B) The percentage of cells counted from Fig. 7A that contain a red prApe1 dot rather than the diffuse red phenotype. Error bars represent the standard deviation from 3 biological replicates. At least 80 cells were counted for each strain. (C) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains under starvation conditions at 30°C. (D) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains under starvation conditions at 37°C. (E) prApe1 processing assay for WT, atg1Δ, arl1Δ and ypt6Δ strains under nonstarvation conditions (time 0), starvation for 3 h at 30°C and starvation for 3 h at 37°C. N, nitrogen. Scale bar: 3 µm.