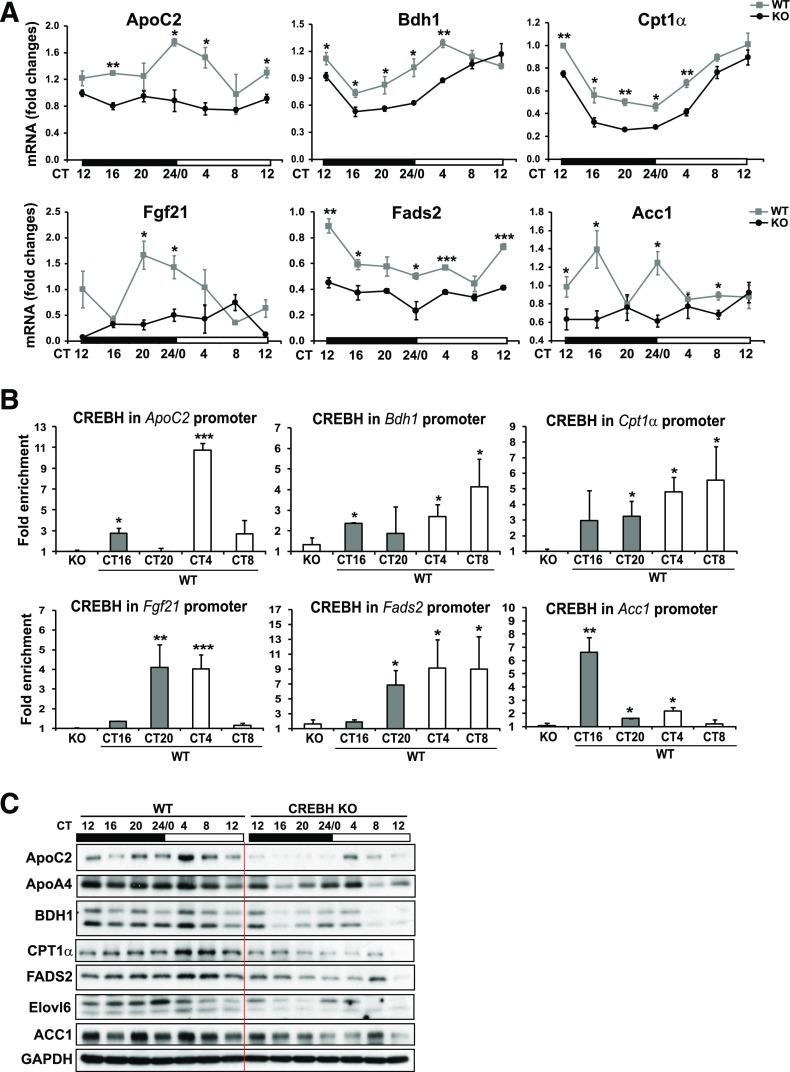
Figure 6.
CREBH regulates expression of the genes involved in lipolysis, FA oxidation, and lipogenesis. A: Rhythmic expression of CREBH target genes involved in lipolysis, FA oxidation, and lipogenesis in CREBH-null and WT mouse livers under the circadian clock. Levels of mRNA were determined by qPCR. Fold changes of mRNA levels were compared with that of one WT mouse at the starting circadian time point. Data are mean ± SEM (n = 3–5 mice/time point). B: CREBH enrichment in CREBH target gene promoters in WT mouse livers during various circadian phases determined by ChIP-qPCR. CREBH-null liver nuclei were used as negative control for the endogenous CREBH ChIP assays. Quantification of CREBH enrichment in the gene promoters at various circadian phases was determined by comparing ChIP-qPCR signals from the samples pulled down by the anti-CREBH antibody with that pulled down by a rabbit anti-IgG antibody. Data are mean ± SEM (n = 3 mice/time point). C: Circadian rhythmic levels of the proteins encoded by the CREBH target genes, including ApoC2, ApoA4, BDH1, CPT1α, FADS2, Elovl6, and ACC1, in the livers of CREBH-null and WT control mice. The liver tissue samples from CREBH-null and WT control mice were collected every 4 h in a 24-h circadian period. Pooled liver protein lysates from three to five mice per genotype group per time point were used for the Western blot analyses. Levels of GAPDH were determined as loading controls. The quantifications of the rhythmic fold changes of these proteins are shown in Supplementary Fig. 4. *P < 0.05, **P < 0.01, ***P < 0.001. KO, knockout.