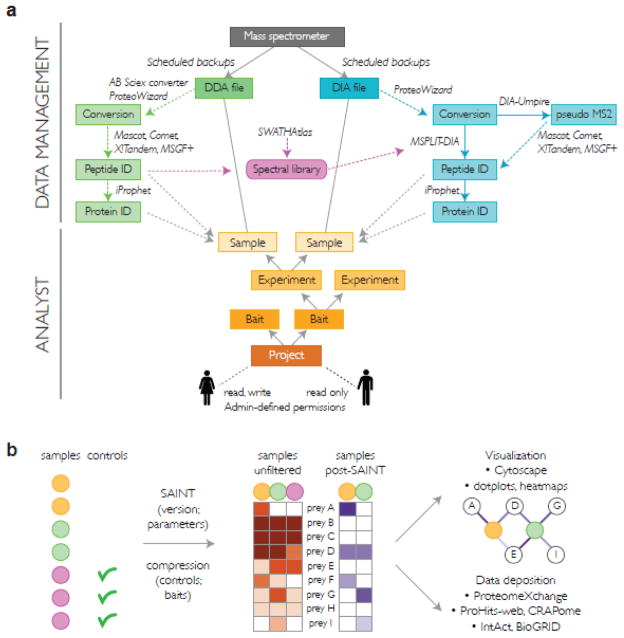
Figure 1. ProHits organization and protein interaction scoring.
A) The ProHits system consists of two principal modules, a Data Management module and an Analyst module. All mass spectrometers in a facility can be connected to ProHits: scheduled backups of the mass spectrometry data are performed followed by file conversion and database searches. DIA identification is supported by DIA-Umpire and the spectral matching tool MSPLIT-DIA. Peptide and protein identification results are parsed to a Sample defined in the Analyst module. The Samples are defined in a Project → Bait → Experiment → Sample hierarchy. Permissions for different projects are assigned to users in an Admin section. B) Schematic workflow for protein interaction analysis using SAINT through ProHits. Within a project, a user defines which samples should be analyzed and specifies which of those are the controls. The SAINT version (SAINTexpress or standard SAINT) is selected, alongside optional parameters and sample compression level. SAINT uses the quantitative matrix to derive the probability of interactions. Post analysis with SAINT, the data can be visualized or deposited in repositories from ProHits itself.