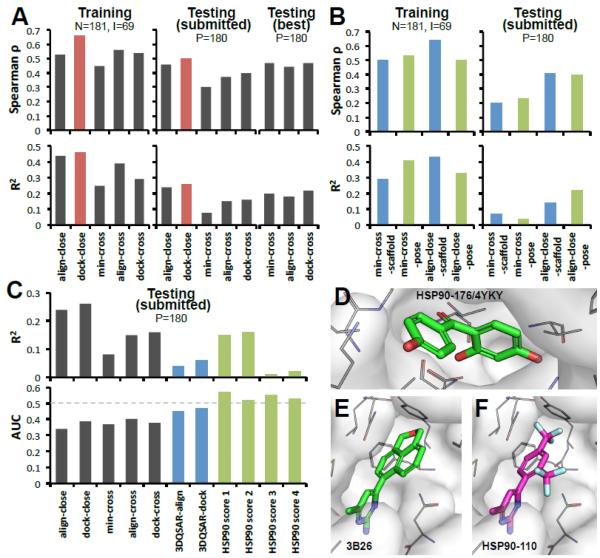
Figure 4. “Close” methods have better performance than “cross” methods for affinity ranking in HSP90 challenge.
A. Prediction rates on: training set, testing set submitted prospectively, and testing (best) set reassessed retrospectively. Optimal receptors for align-cross, min-cross and dock-cross were (prospectively) 3OWD, 4BQJ, 3K98 and (retrospectively) 3T10, 3RLP, 3OWD, respectively. N: number of co-crystals, I: number of co-crystal with IC50 data, P: number of compounds for prediction. B. Results of variant methods: aligning to scaffold, to predicted pose, and using human expertise to eliminate non-binders. C. Distinguishing active from 33 inactive compounds using general methods, human discrimination, 3DQSAR, and special purpose scoring functions to discriminate HSP90 ligands. The lower panel shows binding/non-binding AUC performances, and upper panel shows the corresponding affinity ranking. D-F. Examples of binding poses of inactive compounds. D. Co-crystal of inactive compound 176 (4YKY). E. Co-crystal from PDB 3B26 (unknown IC50). F. prediction for compound 110 (inactive).