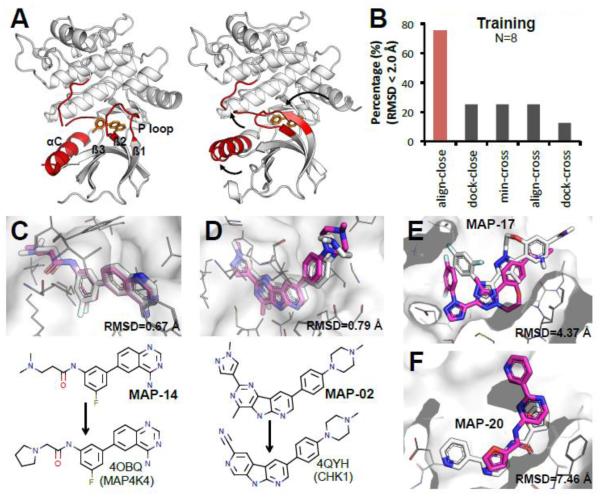
Figure 5. Align-close predicted the best models for 30 MAP4K4 compounds with a mean RMSD of 1.6 Å.
A. Flexible regions around the MAP4K4 binding pocket adapt to different conformations upon ligand binding. (left panel: 4OBO, right panel: 4U44, white cartoon: MAP4K4, red cartoon: flexible loop/helix, orange sticks: small molecules) B. Pose prediction performance across different methods in training set. C, D. Alignment of our best-predicted pose with the co-crystal structure. MAP-14 is an example of aligning to compound from MAP4K4 (RMSD 0.67 Å). MAP-02 is an example of aligning to the compound from other kinase (CHK1, 4QYH) (RMSD 0.79 Å). E, F. Two cases we did poor in pose prediction: MAP-17 and MAP-20. (white sticks: crystal; magenta sticks: predicted; white meshes: MAP4K4 surface; grey lines: MAP4K4 residues close to the binding groove).