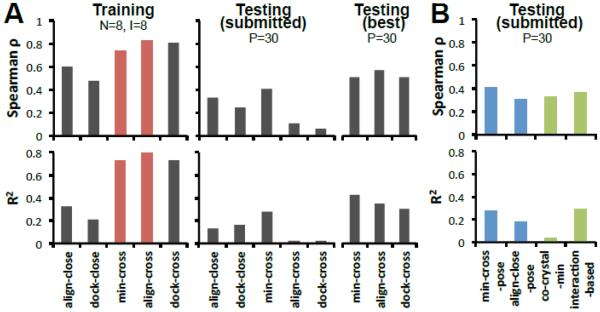
Figure 6. “Cross” methods perform better than “close” methods for affinity ranking of MAP4K4 ligands.
A. Five methods in training set, submitted testing set, and retrospective best predictions for testing set. Optimal receptors for align-cross, min-cross and dock-cross were (prospectively) 4OBP, 4OBP, MAP03 and (retrospectively) MAP29, MAP16, 4U45, respectively. Overall, min-cross and align-cross performed better in our submitted predictions. N: number of co-crystals, I: number of co-crystal with IC50 data, P: number of compounds for prediction. B. Comparison of several pose related methods. (Spearman ρ and R2 are generated by comparing the Vina scores from different methods and experimental IC50 data).