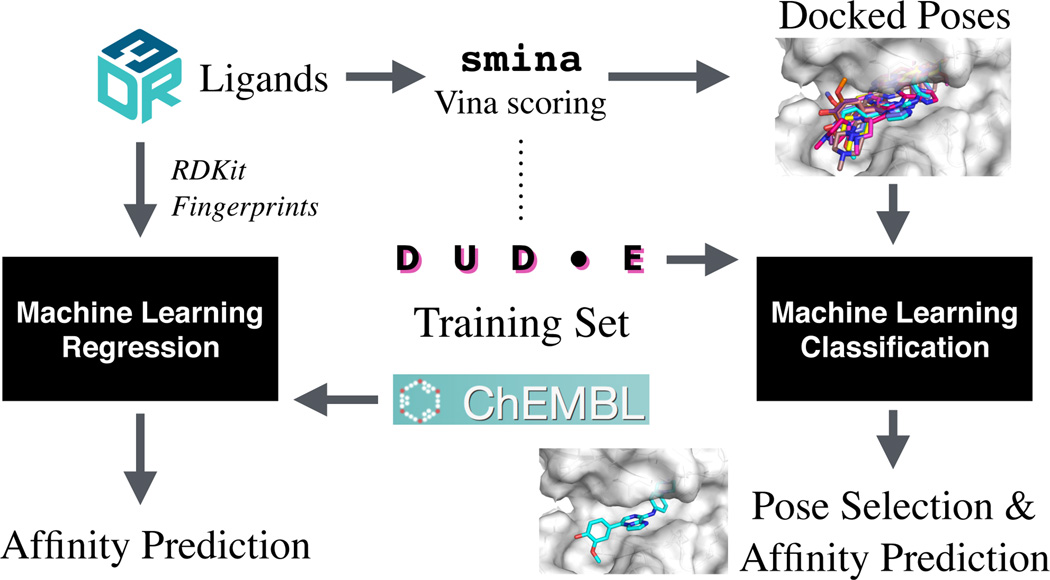
Fig. 1.
The overall approach for our D3R submission. D3R ligands were ranked using a 2D QSAR approach trained using ChEMBL data (left side) or through a structure-based docking and scoring approach that used the DUD-E data set to train custom scoring functions for re-ranking poses docked using smina and the AutoDock Vina scoring function (right side).