Fig. 5.
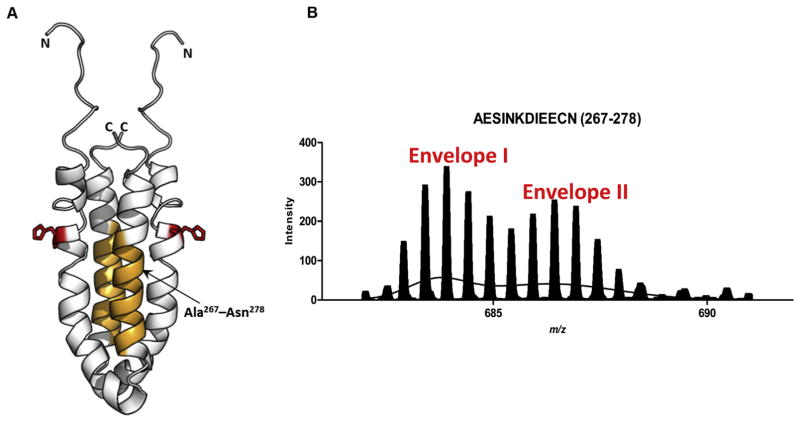
The OmpR binding site of EnvZc displays bimodal Ex1 kinetics (A) Structure of an EnvZc dimer (amino acids 223–289; PDB ID: 1JOY) (Tomomori et al., 1999) is shown in white. Orange residues (267–278) are part of the second helix proximal to the helix containing His-243 (red) and constitute an OmpR-binding site, based on cysteine labeling experiments (Foo et al., 2015), and (Skerker et al., 2008; Szurmant and Hoch, 2010). (B) The spectral envelope of residues 267–278 after 10 min deuterium exchange (charge state: +2) was fit to the sum of two Gaussian distributions. Envelope II corresponded to a significantly higher exchange than Envelope I. Used with permission from Wang et al., 2012.