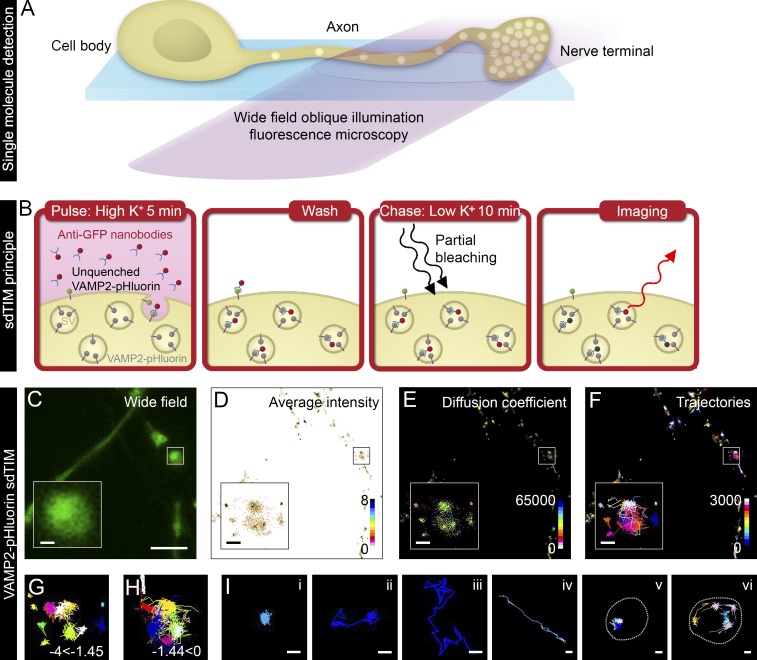
Figure 1.
sdTIM detects the internalized VAMP2–pHluorin–bound Atto647N nanobodies in SVs. (A) Scheme of single fluorescent molecule detection in axons and nerve terminals using oblique illumination. (B) The sdTIM principle: hippocampal neurons expressing VAMP2–pHluorin were stimulated for 5 min with Atto647N nanobody–containing (red) high K+ buffer. After stimulation, the excess nanobodies were washed off, and the neurons were chased for 10 min in low K+ buffer. To detect nanobodies inside individual SVs, the nerve terminals were then exposed to intense laser illumination to bleach the majority of the Atto647N fluorophores, and a subset of internalized Atto647N fluorophores in SVs (red) could then be detected. (C) Wide-field image of hippocampal neuron expressing VAMP2–pHluorin subjected to sdTIM. (D) The corresponding superresolved mean intensity image shows the density map of VAMP2 localization from 3,643 trajectories. The colored bar represents localization densities, and the colder colors indicate higher density. (E) Diffusion coefficient image of VAMP2 localizations from 3,643 trajectories. The detection range from Log10D = −4 to 0 and the warmer colors in the color scale indicate lower mobility. (F) Corresponding trajectories of the VAMP2 mobility over 3,000 frames. Trajectory color-coding refers to acquisition frame number. Boxed areas are shown with higher magnification. Confined (−4 ≤ Log10D ≤ −1.45) (G) and unconfined (−1.45 < Log10D ≤ 0) (H) trajectories of the VAMP2–pHluorin–bound Atto647N nanobodies are shown from the boxed regions indicated in C–F. (I) Representative trajectories of single (i) and dual confinement (ii), free diffusion (iii), directed motion (iv), confined vesicular movement at a site revisited by several vesicles (v; presynapse outlined with dashed line for clarity), and circular movement at the perimeter of an active nerve terminal (vi; presynapse outlined with dashed line for clarity). Bars: (C) 5 µm; (C–F, insets) 0.5 µm; (I) 0.25 µm.