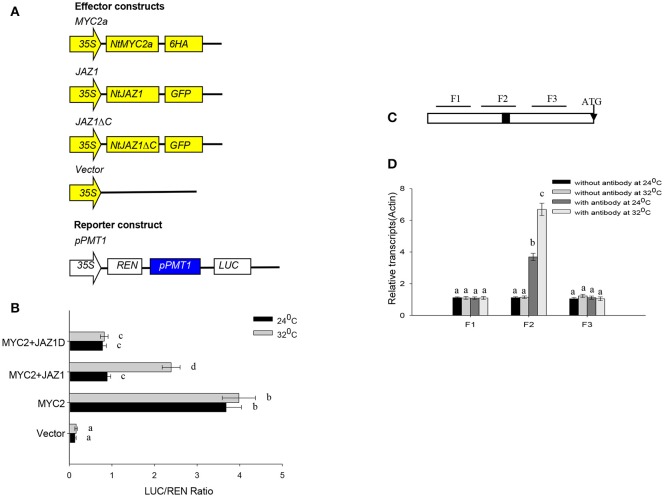
Figure 6.
NtMYC2a binds to the promoter of NtPMT1. (A) Schematic diagrams of the pPMT1 reporter constructs and the NtJAZ1-GFP, NtJAZ1ΔC-GFP lacking the JAS domain.pPMT1 and NtMYC2a empty effector constructs used in the protoplast transcriptional analysis system. (B) Expression assay for the pNtPMT1:LUC in tobacco protoplasts transformed with the NtJAZ1-GFP, NtJAZ1ΔC-GFP or NtMYC2a plasmid or empty vector. The luciferase/REN ratio is shown. Data are the mean ± SD from triplicate experiments. (C) Promoter structure of NtPMT1 gene and the fragment used in the CHIP assay. The black box indicates the G-box motif. Lines indicate the regions (F1, F2, and F3) selected for PCR analysis following ChIP, the detail sequences information and primers sequences for amplifying F1, F2, and F3 fragments show in Supplementary File 2 and Supplementary Table 1. (D) ChIP enrichment showing the ability of MYC2a transcriptional factor to bind in vivo to the promoter region of NtPMT1. ChIP results were normalized to input chromatin, and a fragment in the ACTIN2 promoter was used to normalize the data. Data are the mean ± SD from triplicate experiments. Bars with different letters are significantly difference at p < 0.05 (Tukey's test).