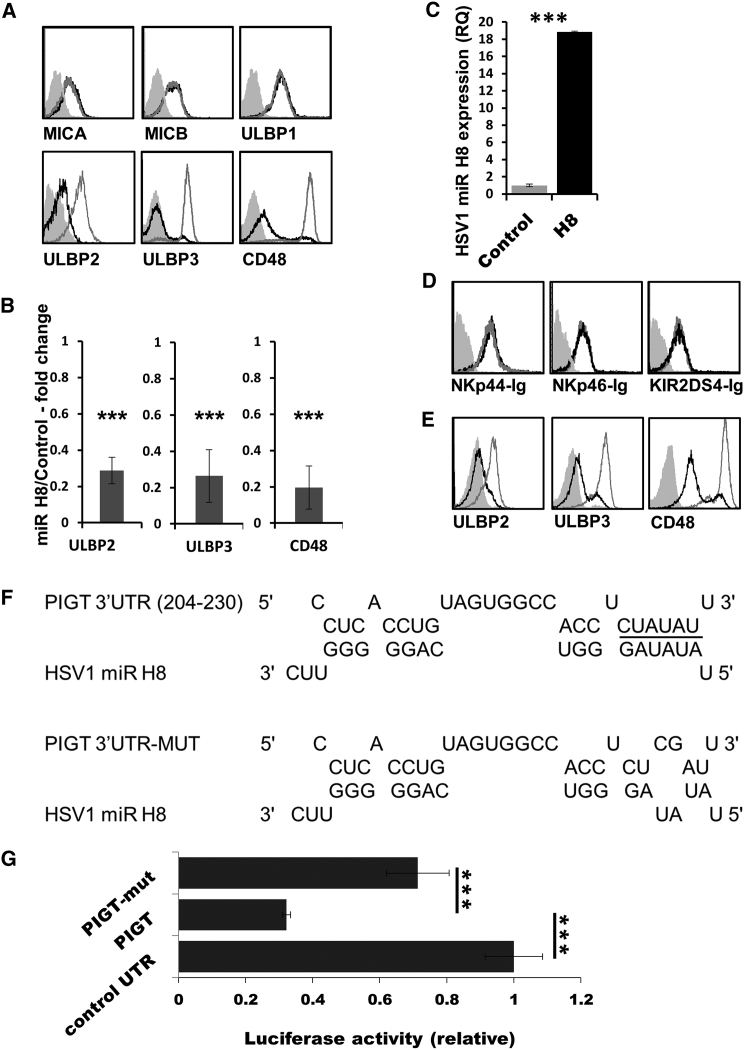
Figure 1.
HSV1 miR H8 Reduces NKG2D and 2B4 Ligand Expression
(A) MiR H8-expressing BJAB cells were stained for the indicated ligands. In all histograms, the black histograms represent the staining of miR H8/or the precursor miR H8, the empty gray histograms represent staining of control SINGFP-expressing cells, and the full gray histograms represent staining of the SINGFP cells with secondary antibody only. The background staining of cells expressing miR H8/pre-miR H8 was similar to that of SINGFP and is not shown.
(B) The fold change in mean fluorescent intensity (MFI) compared with control miRNA-expressing cells was calculated and analyzed using a single sample t test. ULBBP2, n = 5; ULBP3, n = 6; CD48, n = 6. Error bars represent SEM. ∗∗∗p > 0.005.
(C) qRT-PCR for the expression of miR H8. Mean values and SEM error bars are derived from quadruplicates.
(D) BJAB-miR H8/control vector cells were stained with the indicated fusion proteins.
(E) FACS staining of ULBP2, ULBP3, and CD48 in cells expressing pre-miR H8. Staining for all figure parts was repeated at least three times.
(F) Sequence and predicted hybridization of HSV1 miR H8 with its target in the PIGT 3′ UTR (204–230 refers to the base number in the PIGT 3′ UTR). The miR H8 seed region is underlined (top). Shown is sequence alignment of miR H8 and the mutated 3′ UTR of PIGT (bottom).
(G) Relative luciferase activity is displayed for miR H8-expressing HeLa cells (compared with cells expressing a control HSV1 miR H15) transiently transfected with a luciferase reporter attached to a control 3′ UTR, the 3′ UTR of PIGT, or a mutated 3′ UTR of PIGT with a two-base substitution in the seed binding region of miR H8 (as depicted in F).
Shown are mean values and SEM, and error bars were derived from triplicates. The figure shows a representative experiment of four performed. ∗∗∗p > 0.005. See also Figure S1 and S2 and Tables S1 and S2.