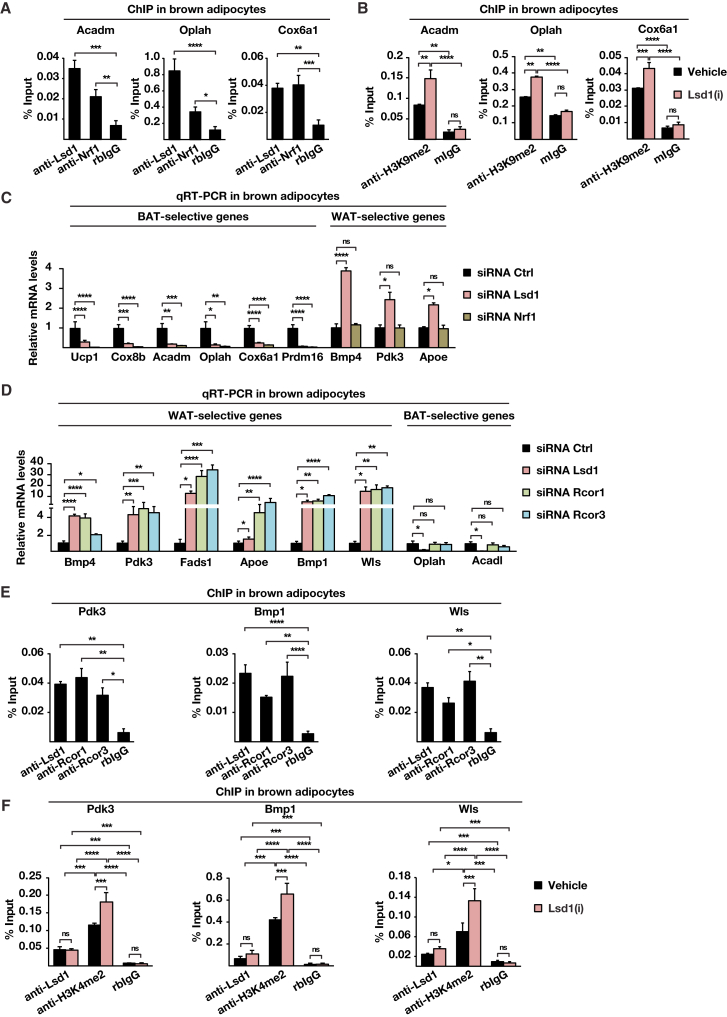
Figure 3.
Lsd1 Regulates the Brown Fat Program through a Dual Mechanism
(A and B) ChIP analysis to detect promoter occupancy performed with anti-Lsd1 and anti-Nrf1 antibodies or rabbit immunoglobulin G (IgG) (rbIgG) in wild-type brown adipocytes (A) and anti-H3K9me2 antibody or mouse IgG (mIgG) in brown adipocytes treated with Lsd1(i) or vehicle (B). The precipitated chromatin was quantified by qPCR analysis with primers flanking Lsd1-binding sites in the indicated genes.
(C and D) Relative mRNA levels of the indicated BAT- or WAT-selective genes in brown adipocytes transfected with unrelated control siRNA (siRNA Ctrl) or siRNA directed against Lsd1 (siRNA Lsd1) or Nrf1 (siRNA Nrf1) (C) and with siRNA Ctrl or siRNA directed against Rcor1 (siRNA Rcor1) or Rcor3 (siRNA Rcor3) (D).
(E and F) ChIP analysis to detect promoter occupancy performed with anti-Lsd1, anti-Rcor1, and anti-Rcor3 antibodies or rbIgG in brown adipocytes (E) and anti-Lsd1 and anti-H3K4me2 antibody or rbIgG in brown adipocytes treated with Lsd1(i) or vehicle (F). The precipitated chromatin was quantified by qPCR analysis with primers flanking Lsd1-binding sites in the indicated genes
Data are mean + SEM. (A), (C), (D), and (E): one-way ANOVA. (B) and (F): two-way ANOVA; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n = 3; qPCRs were run in triplicate. See also Figure S3 and Tables S2, S4, and S5.