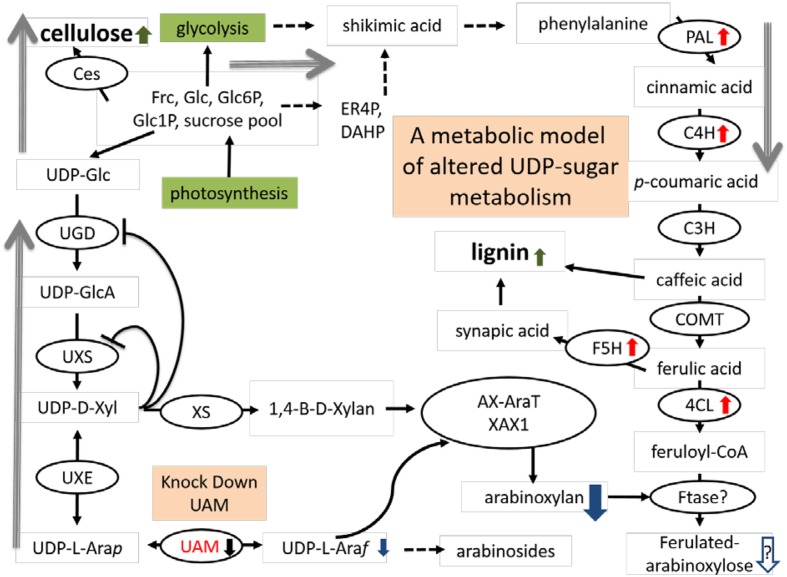
FIGURE 9.
Proposed model arabinoxylan and lignin biosynthesis pathway interactions for downregulated PvUAM1 transgenic switchgrass. Proposed model arabinoxylan and lignin biosynthesis pathway interactions for downregulated PvUAM1 transgenic switchgrass. Biosynthesis proteins denoted in black ovals: Ces, cellulose synthase, UGD, uridine diphosphate (UDP)-glucose dehydrogenase; UXS, UDP-D-xylose synthase, XS, xylan synthase; UXE, UDP-xylose esterase, UAM, UDP-arabinomutase (red lettering); AX-AraT, arabinoxylan arabinosyltransferase (Porchia et al., 2002); XAX1, xylosyl-α-(1,3)-arabinosyl substitution of xylan 1 (Chiniquy et al., 2012); PAL, phenylalanine ammonia-lyase; C4H, coumaroyl shikimate 4-hydroxylase; C3H, coumaroyl shikimate 3-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; F5H, ferulate 5-hydroxylase; 4CL, 4-coumarate: CoA ligase; Ftase?, undetermined feruloyl transferase. Red arrows indicated upregulated genes verified by qRT-PCR. Black arrow indicated downregulation of gene verified by qRT-PCR. Green arrows indicated cell wall components which have been increased. Metabolites are indicated: Frc, fructose; Glc, glucose; Glc6P, glucose-6-phosphate; Glc1P, glucose-1-phosphate; ER4P, erythrose-4-phosphate; DHAP, dihydroxyacetone phosphate; UDP-Glc, UDP-glucose; UDP-GlcA, UDP-glucuronic acid; UDP-D-Xyl, UDP-D-xylose; UDP-L-Arap, UDP-L-arabinopyranose; UDP-L-Araf, UDP-L-arabinofuranose. Blue arrows denote suspected reduction in metabolites resulting from downregulation of PvUAM. Gray arrows indicate shift in flux.