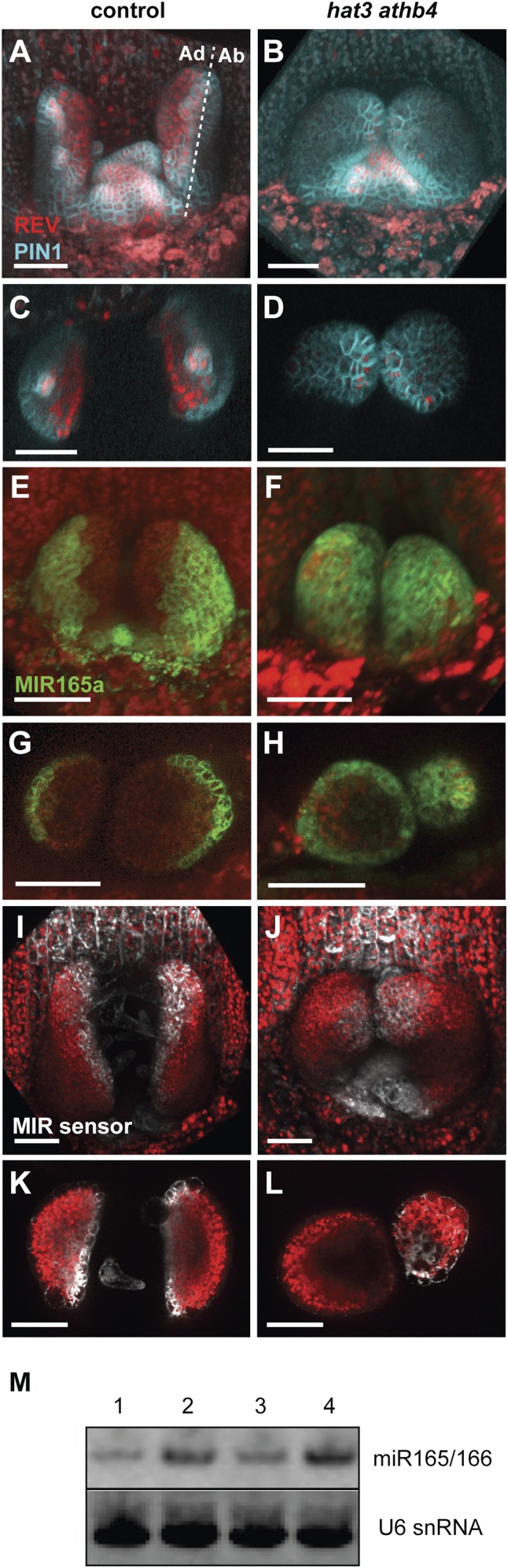
Fig. 1.
HAT3 and ATHB4 are required for repressing MIR165/166 expression. (A–D) Expression of pREV::REV-2xYPET (red) in combination with the auxin efflux carrier PIN-FORMED1 (30, 31) fused to GFP (pPIN::PIN1-GFP) (blue) in the shoot apex of 4-d-old control (A and C) and hat3 athb4 plants (B and D). pPIN::PIN1-GFP is used here to outline the tissue. (C and D) Cross-sections of the same leaf primordia shown in A and B, respectively. (E–H) Expression of pMIR165a::mTagBFP-ER (green) in the shoot apex of 4-d-old control (E and G) and hat3 athb4 plants (F and H). (G and H) Cross-sections of the same leaf primordia shown in E and F, respectively. (I–L) Expression of a miR165/166 sensitive biosensor (white) containing the miRNA target sequence from REV fused to the UV-photoconvertible fluorescent protein mEos2FP (pUBQ10::REV-mEos2FP-ER) in the shoot apex of 4-d-old control (I and K) and hat3 athb4 plants (J and L). The sensitive biosensor is inactivated in cells where miR165/166 are active. (K and L) Cross-sections of the same leaf primordia shown in I and J, respectively. Chlorophyll autofluorescence: red (E–L). (Scale bars, 50 μm.) Ad, adaxial side; Ab, abaxial side. (M) Small RNA Northern blot showing expression levels of miR165/166 and U6 snRNA in Col-0 WT (lane 1), p35S::miR165a (lane 2), hat1 hat2 (lane 3), and hat3 athb4 plants (lane 4).