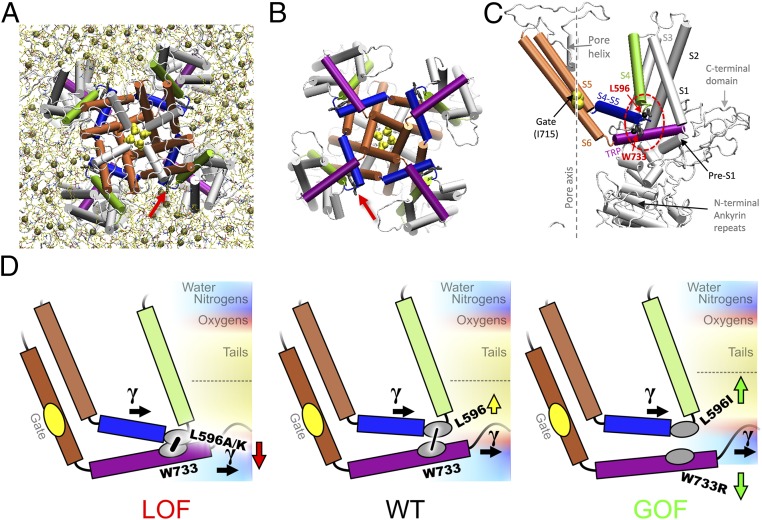
Fig. 1.
Structural elements and the tug-of-war model of TRPV4 gating. A snapshot of the refined TRPV4 homology model (14), viewed from the filter (A) or gate (B) side (cytoplasmic domains not shown). Arrow marks the TRP helix/S4–S5 linker bond. There are about four lipids (tan spheres, phosphorus atoms) per bay in the inner leaflet. (C) In the TRPV4 subunit, S4 (green) is connected to the S4–S5 linker helix (blue) at a sharp elbow contacting the TRP helix (purple). See interactions with lipids in Fig. 5. (D) The expected effect of mutations based on the hydrophobicity of the introduced residue vs. the neighborhood. Mutations at L596 less hydrophobic than leucine are likely to move toward polar regions of the membrane, thus tightening the latch S4–S5 linker/TRP helix contact (low-Po LOF phenotype) (Left), whereas the more hydrophobic isoleucine would be pushed stronger into bilayer core, loosening the latch (GOF phenotype) (Right) and allowing lateral tension (γ) to open the channel. A similar unlatching effect could be caused by the pull on the TRP helix toward the polar region by charged substitutions at W733 (e.g., W733R).