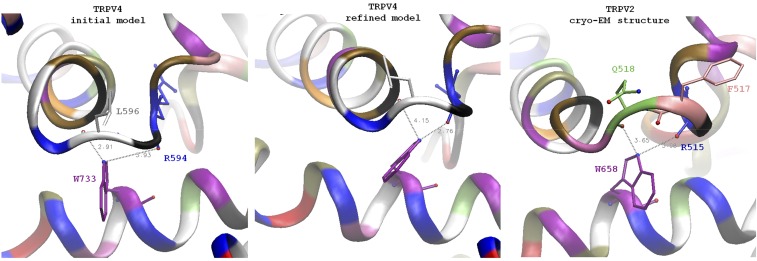
Fig. S5.
Comparison of the TRPV4 original and refined models with the cryoelectron microscopic structure of TRPV2. An overall architecture of the channel in general and of TRP helix contact with S4–S5 linker elbow is similar between the model of TRPV4 and TRPV2 structure. A distinct feature of TRPV2 is a one-residue longer loop between the S4 helix and S4–S5 linker. In TRPV4 the residue K597 is part of S4–S5 linker helix, whereas the residue in the homologous position of TRPV2 (Q518) is not part of the helix and adopts conformation similar to L596. TRPV2 residue F517 (homologous to L596) is in the position intermediate between L596 and G596 of TRPV4. The backbone oxygen of F517 is facing away from the tryptophan and cannot H-bond to it (similar to the backbone in TRPV1 structure with capsaicin). However, the backbone oxygen of Q518 of TRPV2 is in a position similar to the oxygen of L596 of TRPV4 and can potentially serve a similar structural function. Unlike the TRPV4 linker, which has only L596 side chain contacting the lipid bilayer, the linker in TRPV2 has two side chains, F517 and L518, exposed to lipids. This region in TRPV2 might be more tolerant to the point mutations comparing to L596 in TRPV4. The position of tryptophan on the TRP helix is similar to the one in TRPV4: in TRPV2, W658 (homolog of W733) is positioned under the S4–S5 linker elbow. The side chain NH capable of H-bonding is located between the backbone oxygens of Q518 (distance 3.65 Å) and R515 (3.08 Å); note that it is closer to R515 and it agrees with our simulation results on refined models of WT and mutant channels where W733 side chains have switched to R694.