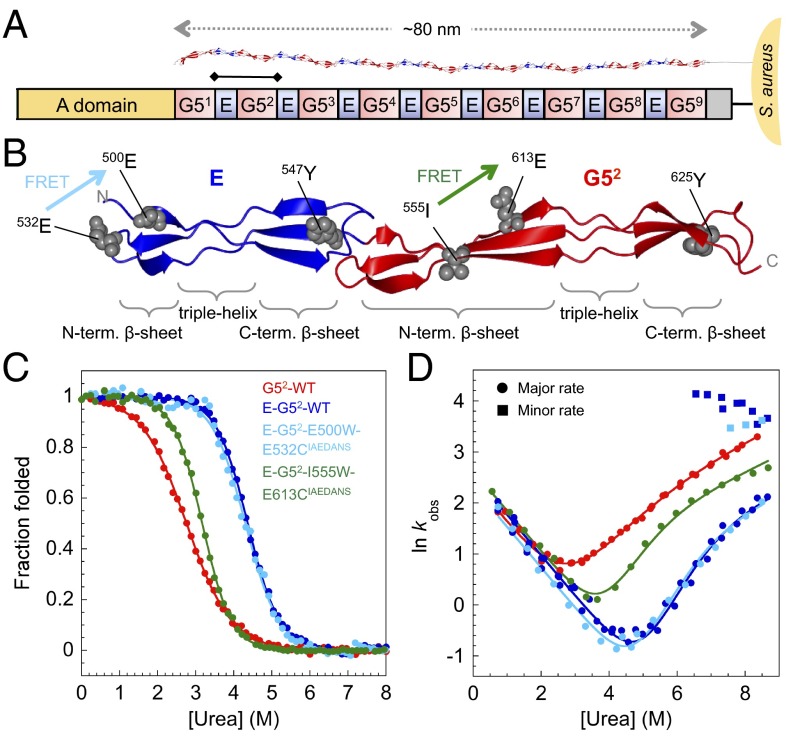
Fig. 1.
Structure and biophysical data for WT SasG G52 and E–G52. (A) Schematic representation of SasG from S. aureus NCTC 8325. The A domain promotes adhesion to host cells. The core region comprises tandemly arrayed G5 (red) and E (blue) domains (10). The E–G52 fragment of SasG is indicated with a bar. (B) Structure of E–G52 (PDB ID code 3TIP) illustrating the topology of E and G52 domains: two single-layer, triple-stranded β-sheets connected by a central collagen-like triple-helical region. The tyrosines and positions of engineered FRET pairs are shown. FRET pair E500W-E532CIAEDANS (cyan) results in FRET only when E is folded; I555W-E613CIAEDANS (green) results in FRET when G52 is folded. (C) Equilibrium denaturation curves. Data for WT G52, E–G52, and E–G52–E500W–E532CIAEDANS were taken from ref. 9. (D) Urea dependence of the natural logarithm of the observed rate constants (in seconds−1) for proteins shown in C. Circles and squares represent major and minor unfolding rate constants, respectively.