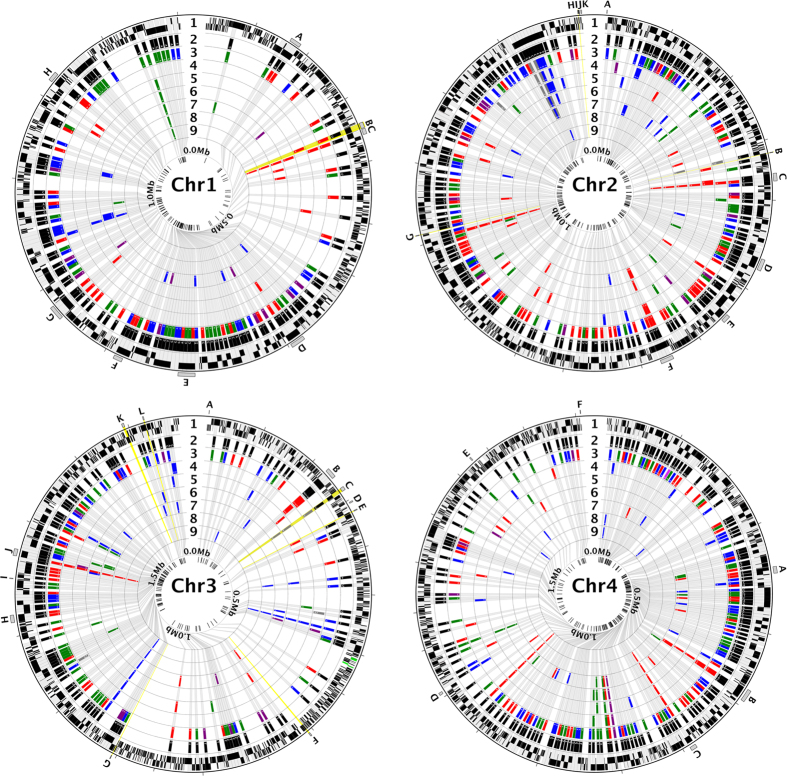
Figure 2. Sequence variants are rare among eight B. microti isolates.
Distribution of all SNPs relative to GI strain, across the variable genes in each of the four nuclear chromosomes of B. microti. In each chromosome plot, the numbered concentric circles represent: 1) genes encoded in the forward and reverse strands (black; rDNA genes in green, in chromosome 3), with loci with sequence variants in any of the strains expanded 5000X; (2) character state, for each variable site, in the GI strain; (3–9) SNPs shown relative to GI, respectively, in R1, Bm1438, ATCC 30222, ATCC PRA-99, GreenwichYale_Lab_Strain_1, Nan_Hs_2011_N11_50, and Naushon. SNP color-code relative to GI: non-synonymous in red; synonymous in green; UTR SNPs in purple; intron and intergenic SNPs in blue. The inner-most circle shows the location of variable loci, with the genomic position shown to scale. The vast majority of SNPs are fixed differences in R1 (track 3), relative to all other strains. Loci that encode the ten most immuno-dominant proteins are highlighted (radial yellow, in chromosomes 1–3; letter-coded). Letter-coded loci also include the 27 most polymorphic genes, which together contain nearly 1/3 of all non-synonymous mutations. Letter code: Chr1: A, BBM_01g00480; B, BBM_01g00985; C, BBM_01g00996; D, BBM_01g01915; E, BBM_01g02065; F, BBM_01g02135; G, BBM_01g02415; H, BBM_01g03070. Chr2: A, BBM_02g00010; B, BBM_02g00896; C, BBM_02g01030 D, BBM_02g01290; E, BBM_02g01490; F, BBM_02g01825; G, BBM_02g03140; H, BBM_02g04250; I, BBM_02g04260; J, BBM_02g04265; K, BBM_02g04280. Chr3: A, BBM_03g00006; B, BBM_03g00690; C, BBM_03g00785; D, BBM_03g00885; E, BBM_03g00947; F, BBM_03g02345; G, BBM_03g03430; H, BBM_03g03985; I, BBM_03g04060; J, BBM_03g04135; K, BBM_03g04695; L, BBM_03g04760. Chr4: A, BBM_04g05770; B, BBM_04g06095; C, BBM_04g06375; D, BBM_04g07535; E, BBM_04g09150; F, BBM_04g09980.