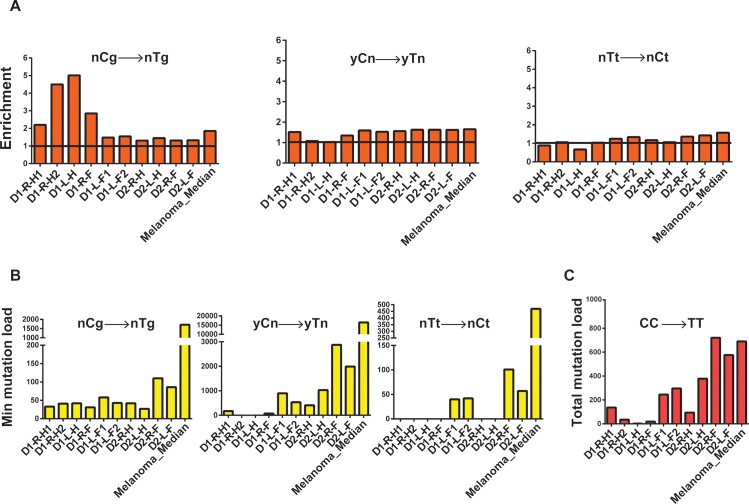
Fig 4. Analysis of mechanistic knowledge-based mutation signatures in the genomes of skin fibroblasts.
Similar analysis for the whole-genome sequenced melanoma (SKCM) cohort (dataset from [46]) is provided for comparison. (A) Fold enrichment of nCg →nTg and UV-signature mutations (yCn→yTn, nTt→nCt; y is either C or T, n is either A, T, G or C, in the trinucleotide context the mutated base is in capital). The black horizontal line denotes the level of no enrichment. (B) The minimum estimates of signature-specific mutation loads for each clone. For the melanoma cohort, the median of the minimum estimated mutation loads for each signature per genome in is shown. (C) Total CC→TT counts of tandem dinucleotide changes in each clone and the median of the total CC→TT counts per genome in the melanoma cohort.