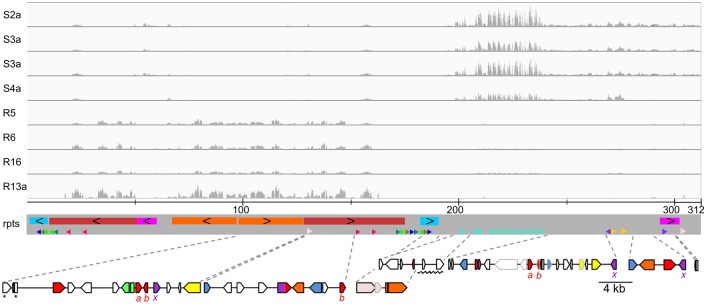
Fig 4. Bipartite structure of chromosome 19.
Genes over-transcribed in virus-resistant lines were all located in a region of ~180 kb (on the left) containing long inverted repeats while under-transcribed genes were on a ~100 kb region (on the right). Tracks from top to bottom: alignment of RNA-Seq reads from susceptible (S2a–S5a) and resistant (R5, R6, R16 and R13a) samples (read depths have the same scale) onto chromosome 19 assembled from PacBio reads; scale in kb; (rpts) rectangles of the same colour on a grey background represent large duplicated blocks with black chevrons indicating inverted relative orientation and small arrowheads and chevrons of the same colour and style represent short repeats and their relative orientation; at the bottom, genomic maps zooming-in on blocks encoding genes under-transcribed (right) and over-transcribed (left) in virus-resistant lines (blue—methyltransferase, red—glycosyltransferase, green—epimerase/dehydratase, purple—sugar phosphate transporter, orange—other sugar metabolism, light pink—cell surface repeat lipoprotein, yellow—transposon-related, white—ORFan or other functions). N.B. the coloured rectangles on the grey background are not related to those used in the gene maps beneath. Significantly differentially transcribed genes are outlined in black (see S2 Table for full annotations of these genes) and those not differentially regulated are outlined in light grey. The gene map region underlined by a zigzag is part of a series of tandem repeats shown by light blue chevrons (see S3 Fig). a, b, x: Genes with the same subscript letter are putative paralogues. *These genes are duplicated as they span the ends of the adjacent inverted repeated blocks but the first gene (ostta19g00040) includes a short tract of unique intervening sequence.