Abstract
Identification of immunoglobulin genes in hybridomas is essential for producing antibodies for research and clinical applications. A couple of methods such as RACE and degenerative PCR have been developed for determination of the Igh and Igl/Igk coding sequences (CDSs) but it has been difficult to process a number of hybridomas both with accuracy and rapidness. Here, we propose a new strategy for antibody sequence determination by mRNA-seq of hybridomas. We demonstrated that hybridomas highly expressed the Igh and Igl/Igk genes and that de novo transcriptome assembly using mRNA-seq data enabled identification of the CDS of both Igh and Igl/Igk accurately. Furthermore, we estimated that only 30,000 sequenced reads are required to identify immunoglobulin sequences from four different hybridoma clones. Thus, our approach would facilitate determining variable CDSs drastically.
Introduction
Hybridomas have been widely accepted as a method for producing large amounts of monoclonal antibodies for research and clinical application [1]. Identification of the amino acid sequence is critical for preserving the characteristics of the antibody, because somatic mutations often occur in the coding region or its regulatory region, resulting in decreased activity of the antibody [2]. Therefore, for the purpose of producing artificial recombinant proteins and filing intellectual properties such as patents, identification of the coding sequences (CDSs) of immunoglobulins is frequently performed to preserve the characteristics of the original antibody.
Antibodies are composed of two subunits; immunoglobulin heavy chain and light chain are each coded by the Igh and Igl/Igk genes. Both subunits have a constant region and a variable region (V region). The constant region is conserved and codes a crystallizable region (Fc region). The V region contains the V, (D) and J segments, and codes the antigen-binding region, also known as Fab region, while Igh has only D segments. These sequences are somatically recombined in pre-B cells, and this recombination plays a key role in antigen specificity and makes it difficult to identify the genomic sequences of immunoglobulin.
A couple of methods have been developed to clone the protein coding sequence of the V region of the Igh and Igl/Igk genes. The 5´RACE method has been widely used to clone the Igh and Igl/Igk sequences from hybridomas [3,4]. However, this method requires a large amount of total RNA. The other convenient method is degenerative PCR, which has also been used, but sometimes it causes loss of the original sequence by mis-hybridization of diverse primers [5–8].
Here, we found that the mRNA-seq data of hybridomas contain a substantial amount of reads derived from Igh and Igl/Igk. De novo transcriptome assembly using whole reads obtained by mRNA-seq enabled us to determine the Igh and Igl/Igk CDSs with only a limited number of reads.
Materials and Methods
Cell lines
The hybridoma cell lines used in this study, 4E5 [Hybridoma clone1 (HD1)], 8H3 [Hybridoma clone2 (HD2)], 5A10 [Hybridoma clone3 (HD3)] and 5F11 [Hybridoma clone4 (HD4)], were generated in our laboratory [9–11]. 4E5 clone was established as previously shown [12]. Mouse hybridoma cell lines, 8A2 and 13C7, producing antibodies against histone H3 Lys9 acetylation, were co-established with CMA310 from the same immunized mouse, as previously described [13,14]. Cells were grown in Hybridoma-Serum Free Media (SFM) made from Hybridoma-SFM powder (Gibco), supplemented with 10% FBS, 1.2% penicillin-streptomycin-glutamine (Gibco) and 1 ng/ml IL-6 or in GIT medium (Wako) containing 1 ng/ml IL-6.
mRNA-seq
Total RNA was extracted from each of the six hybridoma clones (HD1, α-Brg1 antibody, 4E5; HD2, α-Chd2 antibody, 8H3; HD3, α-Chd5 antibody, 5A10; HD4, α-MyoD antibody, 5F11, 8A2, and 13C7) using the AllPrep DNA/RNA Mini Kit (QIAGEN). Library preparation was performed using 1 μg (4E5, 8H3, 5A10 and 5F11) or 3μg (α-Histone H3 lysine 9 acetylatioin (H3K9ac).v2, 8A2 and α-H3K9ac.v3, 13C7) of total RNA and NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs). mRNA-seq was done with an Illumina HiSeq 1500 for 50 bp (4E5, 8H3, 5A10 and 5F11) or 100bp (8A2 and 13C7) paired-end. More than 40M reads were obtained in each sample (HD1 45M reads, HD2 48M reads, HD3 41M reads, HD4 51M reads). We mainly used the mRNA-seq data of HD1 through HD4 and additionally analyzed 8A2 and 13C7 for the comparative study with Sanger sequencing.
mRNA-seq data analysis
The reads were mapped against our custom transcriptome reference sequence, which consists of mouse transcripts (generated from UCSC/mm9 refSeq GTF file in Illumina’s igenome reference set), rat transcripts (generated from the NCBI/Rnor5.0 refSeq GTF file in igenome reference set), and rat Igh and Igl/Igk constant region sequences (obtained from the NCBI nucleotide database; Accession numbers: Ighg1: M28670, Ighg2a: M28669, Ighg2b: M28671, Ighg2c: HQ640952, Iglc1: M22520, Iglc2: M22521, Igk: V01241). BWA-MEM was performed to map sequence reads with the parameter: -t 8 -P -L 10000 (recommended parameter by TIGAR2 [15]). TIGAR2 was run with default settings. The expression level of each gene was quantified as FPKM (fragments per kilobase of exon per million mapped fragments).
De novo transcriptome assembly
Total reads or subsampled reads by fastq-sample (http://homes.cs.washington.edu/~dcjones/fastq-tools) were de novo assembled using Trinity. CPU and max_memory parameters were changed according to each read number (e.g., 40M reads:—CPU 8—max_memory 52G; 1M reads—CPU 2—max_memory 12G). We extracted the Igh and Igl/Igk CDSs by filtering if the contigs contained 20–30 bp of unique sequences of the Igh and Igl/Igk constant region and had proper length (Igh > 1200 bp, Igl/Igk > 600 bp). We developed an automation tool to extract immunoglobulin sequences using Trinity output, which is freely available on http://tx.bioreg.kyushu-u.ac.jp/igfinder.
RT-PCR on the variable region of Igh and Igk
Each hybridoma RNA was obtained by phenol/chloroform extraction. Reverse transcription was performed using the PrimeScript™ II 1st strand cDNA Synthesis Kit (TaKaRa). PCR was performed using the KOD Plus enzyme (TOYOBO) and a thermal cycler. We used the following thermal protocol: predenature at 94°C for 2 min; 35 cycles of 94°C, 15 s, 55°C, 30 s and 68°C, 20 s; final extension at 68°C for 5 min (HD2-Igh, HD3-Igh, HD3-Igk); predenature at 94°C for 2 min; 35 cycles of 94°C, 15 s, 57°C, 30 s and 68°C, 20 s; final extension at 68°C for 5 min (HD1-Igh, HD1-Igk, HD2-Igk, HD4-Igk); predenature at 94°C for 2 min; 35 cycles of 94°C, 15 s, 58°C, 30 s and 68°C, 20 s; final extension at 68°C for 5 min (HD4-Igh).
The HD1-Igh, HD1-Igk and HD4-Igk PCR products were purified by gel extraction, to remove non-specific products. Then, all samples were sequenced by Sanger sequencing.
PCR primers:
HD1-Igh For: 5’-AAAGCATGTGTGTCTGTGATG-3’ (designed on 5ʹUTR region)
HD2-Igh For: 5’-TGAAATCCTCGCAGGAAACTC-3’ (designed on 5´UTR region)
HD3-Igh For: 5’-TGATTGCCACAGCCTTCAGT-3’ (designed on 5´UTR region)
HD4-Igh For: 5’-CATGAAAACCAGCCTGTCCT-3’ (designed on 5´UTR region)
HD-Igh Rev: 5’-AAATAGCCCTTGACCAGGCA-3’ (designed on constant region)
HD1-Igk For: 5’-GAAGGTCTTTCTCAGGGCT-3’ (designed on 5´UTR region)
HD2-Igk For: 5’-GCTCAGCTGTACTCATGC-3’ (designed on 5´UTR region)
HD3-Igk For: 5’-GGTTGGTTGTCATCTTACTGT-3’ (designed on 5´UTR region)
HD4-Igk For: 5’-CTTGTCTTGTTGGCTTGAGAT-3’ (designed on 5´UTR region)
HD-Igk Rev: 5’-TGATGTCTCTGGGATAGAAGTT-3’ (designed on constant region)
Data access
mRNA-seq data were submitted to the DDBJ sequence read archive [DRA004264].
Results
Identification of the hybridoma Igh and Igl/Igk CDSs from mRNA-seq data
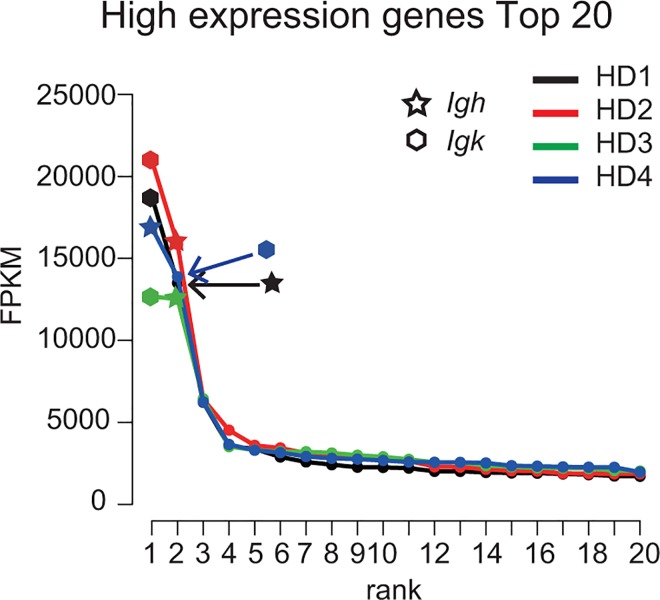
We first performed mRNA-seq on four independent hybridoma clones (HD1, α-Brg1 antibody, 4E5; HD2, α-Chd2 antibody, 8H3 [9]; HD3, α-Chd5 antibody, 5A10 [10]; HD4, α-MyoD antibody, 5F11[11]) that were established as fusion cells of rat B lymphocytes and mouse myeloma cell line SP2 (paired-end 50 bp reads). Then, we comprehensively quantified each transcriptome expression level by BWA-TIGAR2 [15] and ordered them according to expression levels (Fig 1). The data showed that the CDSs of the Igh and Igl/Igk constant region were ranked as the highest expressed transcripts (FPKM > 10000) in all four hybridoma lines (Fig 1). This suggests that the mRNAseq data of hybridomas contained enough number of reads to reconstruct the CDSs of Igh and Igl/Igk [16].
Fig 1. Immunoglobulin gene sequences are highly expressed in all four hybridoma clones.
The graph shows 20 top-ranking gene expression levels in four hybridoma clones HD1–4.The X-axis shows the ranking of the gene expression level in all defined transcripts and the Y-axis shows the expression levels in FPKM. The stars and the hexagons indicate the expression level of the Igh and Igk constant region of the CDS.
Next, we attempted to reconstruct the Igh and Igl/Igk CDSs by de novo transcriptome assembly of the hybridoma mRNA-seq data. First, the mRNA-seq data obtained from hybridoma clone 1 (HD1) were simply assembled with Trinity which reconstructs a full-length transcriptome from RNA-seq data without a genome [16]) without filtering reads (45,406,048 reads), obtaining 58,822 contigs. We further extracted the Igh CDS by filtering if the contigs contained 20–30-bp unique sequences of the Igh constant region, which can uniquely determine each gene (rat-Ighg1: TGTGCCCAGAAACTGTGGAG, rat-Ighg2a: GCCAAGGGAATGCAATCCTTG, rat-Ighg2b: CAAACAACAGCCCCATCTGTCTAT, rat-Ighg2c: AGAACAACAGCCCCATCTGTCTA). As the full length of IgH has more than 400 amino acids (aa) [17], a 1395-bp sequence was obtained as the Igh CDS containing the unique 24-bp sequence of Ighg2b after filtering if the contig has more than 1200 bp. The V region and partial constant region of the obtained Igh CDS were confirmed to be identical to the sequence obtained by Sanger sequencing of reverse-transcribed PCR (RT-PCR) products which were amplified with primer sets designed based on the 5'UTR region and constant region. Alignment of the obtained IgH protein sequence and the known rat IgH constant region (AAA60738) confirmed matching of the full length of amino acid sequence of the constant region in the known IgG2b and the 133–464 amino acids identified in the Igh CDS (Fig 2B). We also extracted the Igl/Igk CDS from the contigs by filtering if it contains 20–30-bp unique sequences of the known Igl/Igk constant region (rat-Igl1: CAACCCAAGGCTACGCCCTC, rat-Igl2: CAGCCCAAGTCCACTCCCAC, rat-Igk: ACCAACTGTATCTATCTTCCCACCATCCAC). As the full length of IgK has more than 200 aa [17], a 705-bp sequence was obtained as the Igk CDS after filtering if the contig has more than 600 bp (Fig 2C). We also confirmed the V region and partial constant region of the obtained Igk CDS with Sanger sequencing following RT-PCR. We demonstrated matching of the amino acid sequence of the constant region in known IgK (CAA24558) and the amino acids of the identified Igk CDS (Fig 2D). We also identified the Igh and Igl/Igk CDSs of HD2 (Ighg2a /Igk), HD3 (Ighg2a /Igk) and HD4 (Ighg2a /Igk) (data not shown). Then, these identified antibody isotypes corresponded to the results of ELISA-format isotyping assay. Mouse Igh and Igk CDSs from mouse hybridoma clones (8A2, 13C7) were also identified and their amino acids sequences were identical to coding sequence determined by PCR cloning shown in [7,13] except the regions coded on sequences on degenerative sequences (S1 Fig). Mouse Igh and Igl/Igk transcripts was extracted by unique sequences of the mouse Igh and Igl/Igk constant region (mouse-Ighg1: CCAAAACGACACCCCCATCT, mouse-Ighg2a: GTGTGTGGAGATACAACTGGCT, mouse-Ighg2b: CCAAAACAACACCCCCATCAG, mouse-Ighg2c: GTGTGGAGGTACAACTGGCTCCT, mouse-Ighg3: CTACAACAACAGCCCCATCTG, mouse-Igl1: GCCAGCCCAAGTCTTCGCCAT, mouse-Igl2: GTCAGCCCAAGTCCACTCCCACTC, mouse-Igl3: GTCAGCCCAAGTCCACTCCCACAC, mouse-Igl4: GCCAACCCAAGGCTACACCCTCAG, mouse-Igk: GGGCTGATGCTGCACCAACTG). These results indicated that simple de novo assembly using hybridoma mRNA-seq data was beneficial for identifying both the Igh and Igl/Igk genes.
Fig 2. Identified immunoglobulin nucleotide and protein sequences of HD1.
(A) The partial nucleotide sequence of Igh and the corresponding amino acid sequence. (B) Sequence alignment of HD1 IgH protein (top lane) and the known rat IgH constant region (bottom lane). The Igh CDS of HD1 codes the amino acid sequence of the Ig gamma-2B chain C region. (C) The partial nucleotide sequence of Igk and the amino acid sequence. The Igh CDS and Igk CDS were obtained after de novo assembly of HD1 mRNA-seq data and filtering contigs. (D) Alignment of the IgK protein sequence of HD1 and the known rat IgK constant region. The Igk CDS of HD1 codes the amino acids of the Ig kappa chain C region. White, red and blue backgrounds show the UTR region, V region and C region, respectively.
Optimization of de novo transcriptome assembly for identifying immunoglobulin sequences
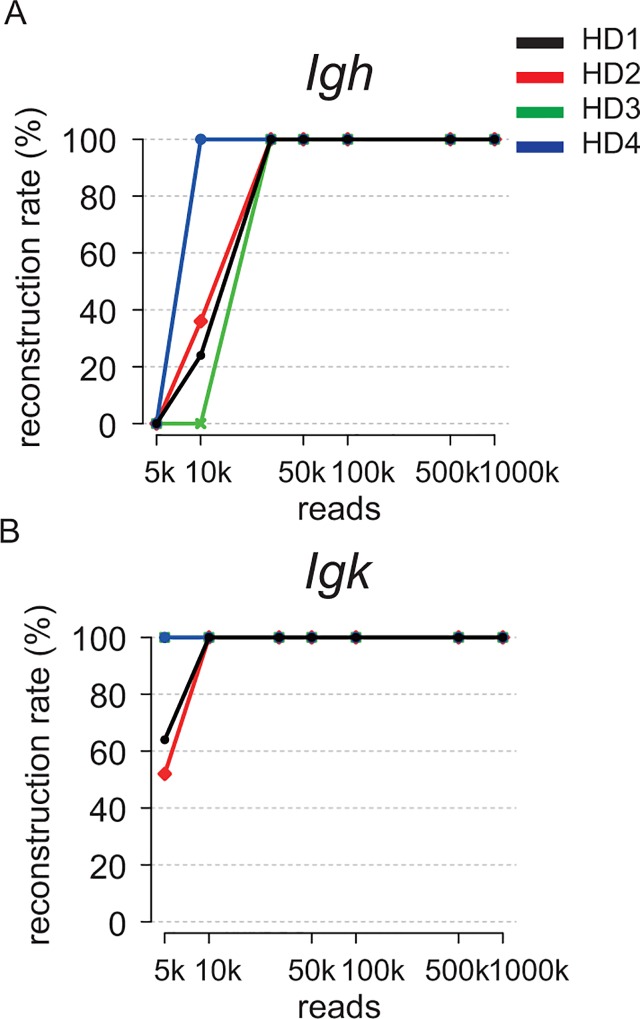
We further attempted to optimize the strategy for identifying the CDSs of the Igh and Igl/Igk genes using hybridoma mRNA-seq data. First, to estimate the required number of reads for identifying an immunoglobulin gene by our method, we randomly subsampled 5k, 10k, 30k, 50k, 100k, 500k and 1M reads from the total reads in the mRNA-seq data of four different hybridomas (HD1, HD2, HD3 and HD4). Then, we repeated the de novo assembly 25 times using the randomly selected reads. We defined the Igh and Igk CDSs identified by the de novo assembly using the total reads (such as in Fig 2A and 2C) as correct sequences, and then calculated the success rate of obtaining complete CDSs (Fig 3A and 3B). The Igh and Igk CDSs of all four clones were perfectly identified with > 30k reads (Fig 3A) and > 10k (Fig 3B) reads, respectively. This result confirmed that our method successfully identified immunoglobulin sequences with limited reads from mRNA-seq data (Fig 4).
Fig 3. Required number of reads to identify immunoglobulin gene sequences was only 10–30k.
The number of reads used for de novo assembly and the success rate (%) of obtaining complete Igh CDS (A) and Igk CDS (B). Immunoglobulin sequences from four hybridomas were assembled with 30k reads. The random selection of reads and de novo assembly were performed 25 times for each number of subsampled reads.
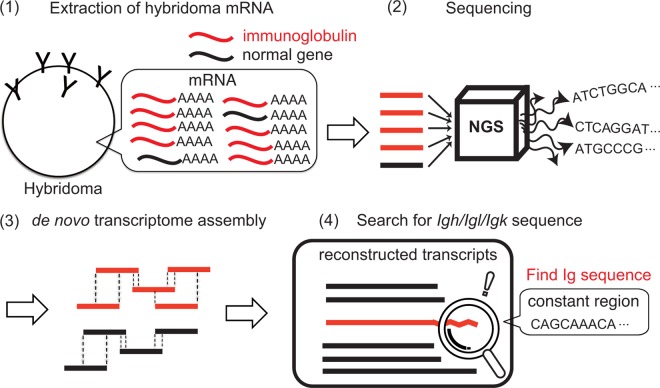
Fig 4. Schematic drawing of the immunoglobulin gene sequences identification using a low read number of hybridoma mRNA-seq data.
First, mRNA-seq of hybridoma is performed (step 1, 2). Next, mRNA-seq data is de novo assembled (step 3). Finally, promising immunoglobulin gene sequences are identified by filtering contigs according to the requirements for coding valid immunoglobulin protein sequences (step 4).
We implemented our immunoglobulin sequence identification strategy with a python script named igfinder available at our website (http://tx.bioreg.kyushu-u.ac.jp/igfinder).
Discussion
Here, we propose a rapid and accurate method for identifying the CDSs of Igh and Igl/Igk by de novo transcriptome assembly. Our method requires limited reads of mRNA-seq, because hybridomas highly express Igh and Igl/Igk transcripts. Our approach would be beneficial for rapid and cost-effective cloning of Igh and Igl/Igk CDSs.
Conventionally, PCR, 5'RACE and SMARTer RACE (Clontech) have been used with degenerative primers for the determination of antibody sequences (S1 Table). 5´RACE has been widely used to identify Igh and Igl/Igk CDSs from hybridomas; however, it is time consuming and requires a large amount of total RNA. SMARTer RACE, which is a refinement on 5'RACE, requires only a small amount of RNA; however, SMARTer RACE and 5’RACE occasionally extract pseudo-sequences caused by annealing or mis-annealing of primers to the myeloma cell-derived Igh or Igl/Igk sequences in the hybridoma [3]. Therefore, several clones’ identified sequences should be confirmed by other approaches such as Sanger sequencing of RT-PCR products. Our method avoided this procedure, because the Igh and Igl/Igk sequences were selected by filtering sequences based on the CDS length. We also surmise, on account of the remarkably high expression of Ig genes, that our method can work with as low as ~0.1μg of total RNA, which is the minimal requirement for the library prep kit used in this study.
Our method depends on the quantity of Igh and Igl/Igk transcripts in each hybridoma. Therefore, hybridomas that express antibodies with a low level of Igh and Igl/Igk may not have enough transcripts to be assembled. In this case, increasing the read number for de novo assembly could be beneficial for identifying the Igh and Igl/Igk CDSs [16].
Another advantage of our method is the ability to identify full-length Igh and Igl/Igk, while other methods only identify the V region of immunoglobulin genes. Therefore, our method enables identification of the antibody isotypes and subclasses (e.g., rat- Ighg1, Ighg2a, Ighg2b, Ighg2c). We hope to extend our method in our future work for the detection of minor variants of antibody genes caused by somatic mutations, e.g., in clinical samples of myeloma or lymphoma cells.
A recent study has demonstrated that mouse plasma cells highly express immunoglobulin genes [18]. Hybridomas also highly express immunoglobulin genes, which are derived from a B cell fused with a myeloma cell. Our data suggest that it is possible to identify Igh and Igl/Igk CDSs, even from intact B cells or plasma cells, even at the single cell level.
Supporting Information
(EPS)
(PDF)
Acknowledgments
We thank A. N. Imbalzano, T. Tachibana for insightful discussions; T. Ichinose, M. Kato, N. Ikeda, S. Hirata, M. Mouri, M. Harada for technical support; and “Advanced Computational Scientific Program” of Research Institute for Information Technology, Kyushu University and the National Institute of Genetics (NIG) for providing the high-performance computing resources.
Data Availability
All mRNA-seq files are available from the DDBJ database (accession number DRA004264).
Funding Statement
This work was supported by the Core Research for Evolutional Science and Technology (CREST) (http://www.jst.go.jp/kisoken/crest/) and open access charge: Japan Society for the Promotion of Science (JSPS) KAKENHI [grant numbers 23310134, 25116010, 25132709, 25118518, 26290064] (https://www.jsps.go.jp). We have had the support from the updated grants (JSPS KAKENHI: 15K18457, 16H01219, 16K18479, 16H01577, 16H01550). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gura T. Therapeutic antibodies: magic bullets hit the target. Nature. 2002. June 6;417(6889):663–73. [DOI] [PubMed] [Google Scholar]
- 2.Xin H and Cutler J.E. Hybridoma passage in vitro may result in reduced ability of antimannan antibody to protect against disseminated candidiasis. Infect Immun. 2006. July;74(7):4310–21. 10.1128/IAI.00234-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruberti F, Cattaneo A and Bradbury A. The use of the RACE method to clone hybridoma cDNA when V region primers fail. J Immunol Methods. 1994. July 12;173(1):33–9. [DOI] [PubMed] [Google Scholar]
- 4.Doenecke A, Winnacker E.L and Hallek M. Rapid amplification of cDNA ends (RACE) improves the PCR-based isolation of immunoglobulin variable region genes from murine and human lymphoma cells and cell lines. Leukemia. 1997. October;11(10):1787–92. [DOI] [PubMed] [Google Scholar]
- 5.Dubel S, Breitling F, Fuchs P, Zewe M, Gotter S, Welschof M, et al. Isolation of IgG antibody Fv-DNA from various mouse and rat hybridoma cell lines using the polymerase chain reaction with a simple set of primers. J Immunol Methods. 1994. September 30;175(1):89–95. [DOI] [PubMed] [Google Scholar]
- 6.Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard H.R., et al. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J Immunol Methods. 1997. February 14;201(1):35–55. [DOI] [PubMed] [Google Scholar]
- 7.Zhou H, Fisher R.J and Papas T.S. Optimization of primer sequences for mouse scFv repertoire display library construction. Nucleic Acids Res. 1994. March 11;22(5):888–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honegger A. and Plückthun A. The influence of the buried glutamine or glutamate residue in position 6 on the structure of immunoglobulin variable domains. J Mol Biol. 2001. June 8;309(3):687–99. 10.1006/jmbi.2001.4664 [DOI] [PubMed] [Google Scholar]
- 9.Harada A, Yoshimura S, Odawara J, Azuma M, Okada S, Nakamura M, et al. Generation of a rat monoclonal antibody specific for CHD2. Hybridoma (Larchmt). 2010. April;29(2):173–7. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura S, Yoshimi T, Ohkawa Y, Azuma M and Tachibana T. A rat monoclonal antibody against the chromatin remodeling factor CHD5. Hybridoma (Larchmt). 2010. February;29(1):63–6. [DOI] [PubMed] [Google Scholar]
- 11.Harada A, Ohkawa Y, Ao S, Odawara J, Okada S, Azuma M, et al. Rat monoclonal antibody specific for MyoD. Hybridoma (Larchmt). 2010. June;29(3):255–8. [DOI] [PubMed] [Google Scholar]
- 12.Ohkawa Y, Harada A, Nakamura M, Yoshimura S and Tachibana T. Production of a rat monoclonal antibody against Brg1. Hybridoma (Larchmt). 2009. December;28(6):463–6. [DOI] [PubMed] [Google Scholar]
- 13.Sato Y, Mukai M, Ueda J, Muraki M, Stasevich TJ, Horikoshi N, et al. Genetically encoded system to track histone modification in vivo. Sci Rep. 2013;3:2436 10.1038/srep02436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011. August;39(15):6475–88. 10.1093/nar/gkr343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nariai N, Kojima K, Mimori T, Sato Y, Kawai Y, Yamaguchi-Kabata Y, et al. TIGAR2: sensitive and accurate estimation of transcript isoform expression with longer RNA-Seq reads. BMC Genomics. 2014;15 Suppl 10:S5 10.1186/1471-2164-15-S10-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabherr M.G, Haas B.J, Yassour M, Levin J.Z, Thompson D.A, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011. May 15;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elgert K.D. Antibody Structure and Function. In Immunology: understanding the immune system Wiley-Liss; 1996; pp. 58–78. [Google Scholar]
- 18.Shi W, Liao Y, Willis S.N, Taubenheim N, Inouye M, Tarlinton D.M, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015. June;16(6):663–73. 10.1038/ni.3154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(EPS)
(PDF)
Data Availability Statement
All mRNA-seq files are available from the DDBJ database (accession number DRA004264).