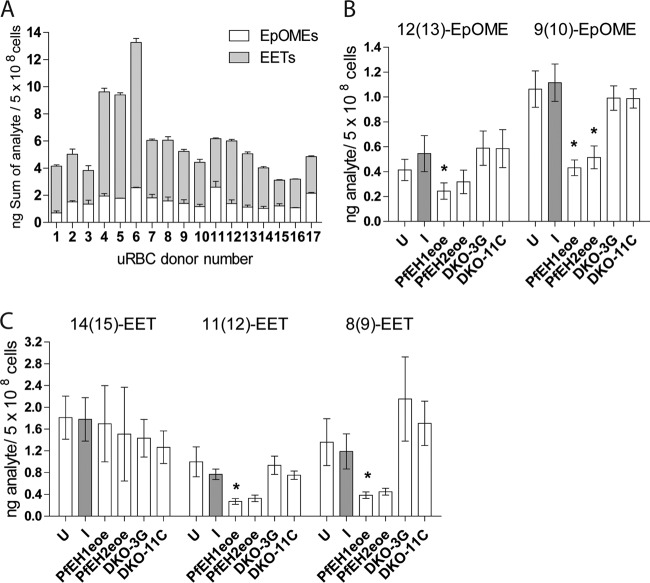
FIG 6 .
LC/MS/MS measurement of epoxide-containing fatty acids in phospholipid preparations from parasitized RBCs. Internal standards 5(6)-EET-d11, 8(9)-EET-d11, 11(12)-EET-d11, 14(15)-EET-d11, and 9(10)-EpOME-d4 were added to each sample, phospholipids were extracted with 2:1 CHCl3−CH3OH plus 0.1 mM triphenylphosphine, and fatty acids were released using phospholipase A2. Four-point calibration curves were generated using the deuterated standards to quantify the epoxide analytes. (A) Sum of 8(9)-EET, 11(12)-EET, 14(15)-EET, 9(10)-EpOME, and 12(13)-EpOME in uninfected RBCs (uRBC). Samples were run twice, and the error bars depict range/2. (B and C) 9(10)-EpOME and 12(13)-EpOME (B) and 8(9)-EET, 11(12)-EET, and 14(15)-EET (C) concentrations in phospholipids of uninfected RBCs (U) and RBCs infected with 3D7 parent (I), PfEH1-GFP (PfEH1eoe), PfEH2-GFP (PfEH2eoe), and PfEH1 PfEH2 double-knockout (DKO) clones 3G and 11C. The values shown are calculated from 3 to 9 independent phospholipid preparations per condition (mean ± SEM). Values that are statistically significantly different from the value for infected RBCs (P < 0.05 by paired t tests) are indicated by an asterisk. For PfEH1, there was a significant reduction in 12(13)-EpOME (P = 0.049; n = 4), 9(10)-EpOME (P = 0.043; n = 4), 11(12)-EET (P = 0.0154; n = 4), and 8(9)-EET (P = 0.034; n = 4), while for PfEH2, several compounds showed reductions, but the decrease reached statistical significance only for the 9(10)-EpOME analyte (P = 0.028; n = 4).