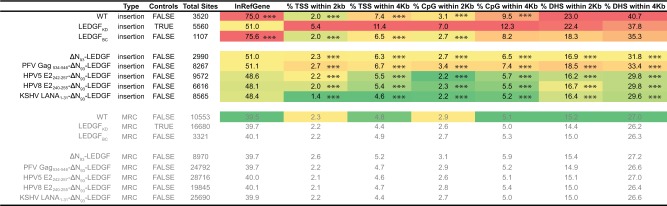
Fig 5. Integration frequency near genomic features.
Fig showing the percentage HIV-derived lentiviral vector integration sites relative to features specific for integration viral vectors such as integration into the body of genes (Refseq genes, InRefGene), integration within 2kb-4kb windows near Transcription Start Sites (X5-end of genes, TSS), midpoint of CpG islands or DNase I-hypersensitive sites (DHS), counted in both the 5’ and 3’ direction. Dataset details are described in the MM section. Asterisks depict a significant deviation from LEDGF KD (two-tailed Chi-square test; ***, p-values <0.001). TSS, Transcription start sites; DHS, DNase I-hypersensitive sites; PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen; MRC, matched random control.