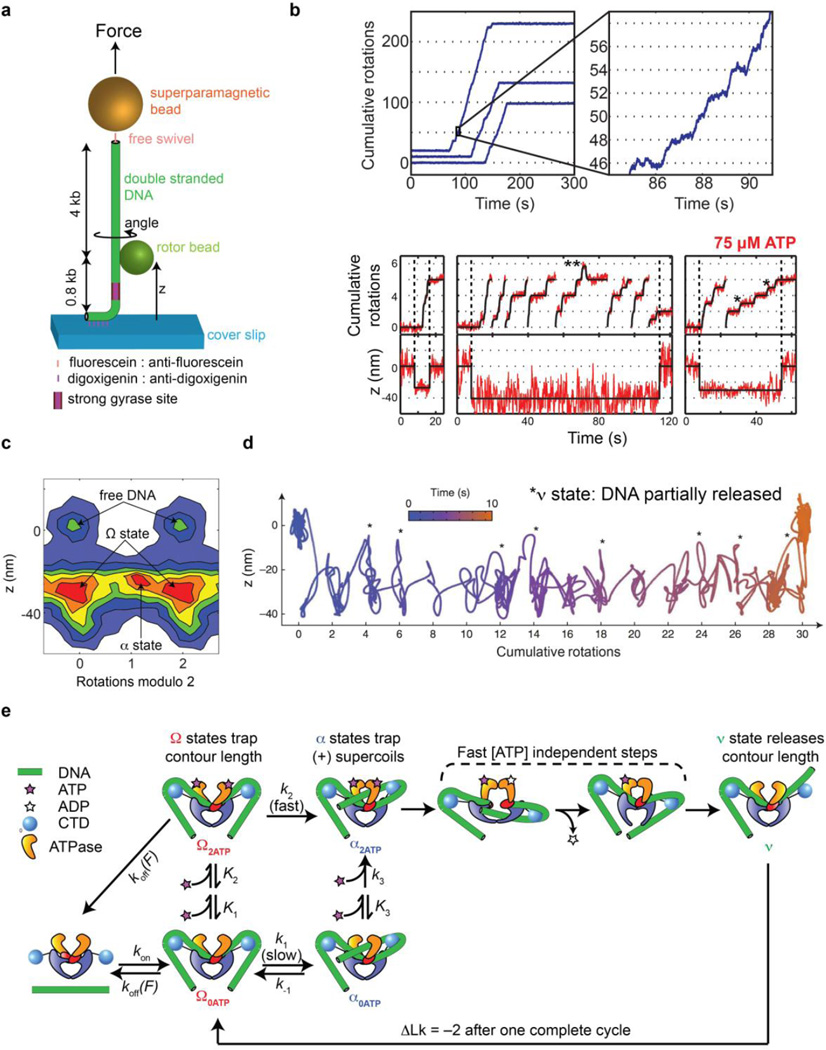
Figure 4.
Rotor bead tracking reveals new conformations and ATP-dependent dynamics of E. coli DNA gyrase. (a) The rotor bead tracking (RBT) assay. DNA is stretched using a magnetic bead, and a submicron rotor bead is attached to the side of the molecule and tracked using fluorescence [35] [36] or evanescent scattering [37] videomicroscopy to measure changes in DNA angle and extension (z) in real time. (b) RBT traces (reproduced from [36]) in the presence of DNA gyrase under in 1 mM ATP (above) or 75 µM ATP (below). Individual gyrase encounters lead to bursts of stepwise rotation, corresponding to processive negative supercoiling. [ATP]-dependent dwells are seen at the even rotation mark and also at an intermediate angle (*) corresponding to a chirally wrapped intermediate. (c) 2D histogram (reproduced from [36]) of paired (angle,z) values during gyrase activity in presence of 75 µM ATP, showing distinct conformational states visited by the enzyme. Angles are shown modulo 2 rotations. The Ω state is significantly contracted in z but lies at the ~0 rotation mark, which is explained by sequestering DNA contour without trapping writhe. The α state can also be seen at the ~1 rotation mark, corresponding to trapping (+) writhe prior to strand passage. (d) High-resolution dynamics of gyrase at 1 mM ATP using gold rotor bead tracking (reproduced from [37]). A single processive burst is shown in the (angle, z) plane. Major dwells are interrupted by brief excursions to a state (*) that releases significant contour length. (e) Branched kinetic model for structural transitions and ATP coupling in DNA gyrase [36, 37]. The kinetics of processive supercoiling are dominated by the transition from Ω to α, which can occur slowly and reversibly in the absence of ATP or quickly when 2 ATP are bound. Subsequent strand passage also requires the presence of 2 ATP. DNA is partially released after strand passage and recaptured to begin a new round of supercoiling.