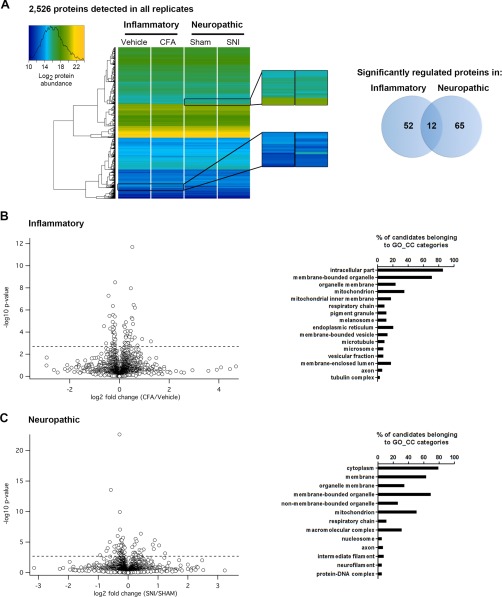
Fig. 2.
Results of DIA-MS proteomics of inflammatory and neuropathic pain in mice. A, Left, Heatmap upon unsupervised hierarchical clustering analysis of abundance levels of the same set of 2526 proteins quantified in all four experimental groups. Magnified regions show examples of changes in abundance (represented as changes in the color code) of specific proteins. Right, Venn diagram shows the number of specifically and commonly regulated proteins (intersection) detected in inflammatory and neuropathic pain models, respectively. B, Left, Volcano plot for all proteins profiled in inflammatory (CFA/Veh) pain. Dotted horizontal line represents the significance level, i.e. q-value = 0.05. Hence, all proteins above this dotted line exhibit a q-value < 0.05 and are considered to be significantly regulated. Right, Assigned Gene Ontology (GO) Cellular Component (CC) categories and the relative abundance of significantly regulated proteins during inflammatory pain. C, Left, Volcano plot for all proteins profiled in neuropathic (SNI/Sham) pain. Significance level q-value < 0.05 as in B. Right, Assigned Gene Ontology (GO) Cellular Component (CC) categories and the relative abundance of significantly regulated proteins during neuropathic pain.