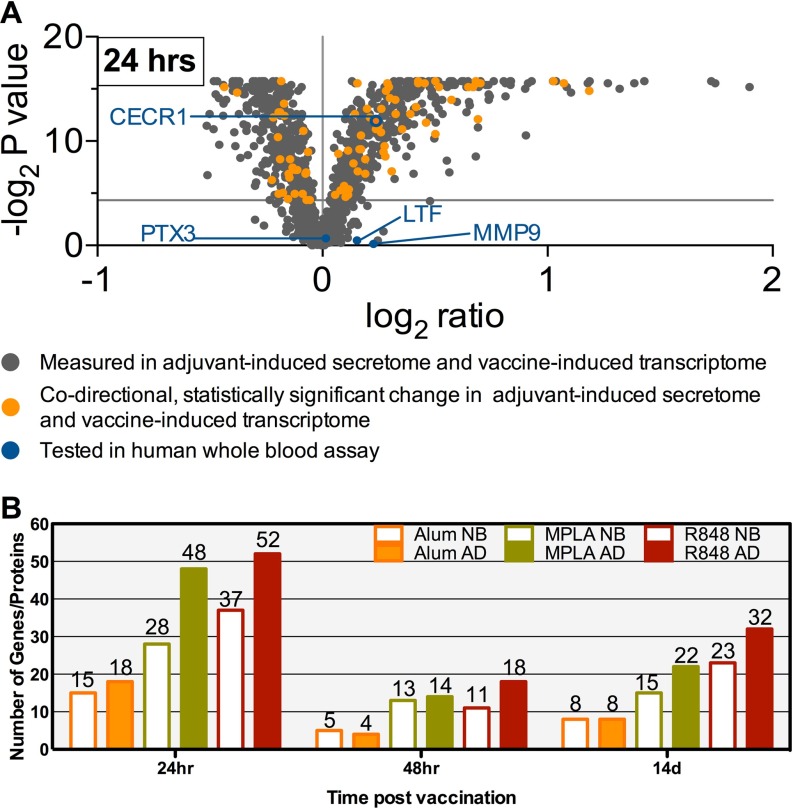
Fig. 7.
Gene Expression Levels in Adults Immunized with an MPLA-Adjuvanted Malaria Vaccine Demonstrate Adjuvant-, Age- and Kinetic-Concordance with Secretome Proteins in Silico. PBMC transcriptome profiles of 24 adults immunized with the MPLA-aduvanted Malaria vaccines RTS,S/AS01B or RTS,S/AS02A, measured at four different time points (49) were analyzed for post-immunization changes in gene expression. A, For each protein identified in the adjuvant-induced monocyte secretomes, the corresponding gene's fold change and p value at the 24 h time point was plotted on the indicated log scales. Highlighted in orange are those molecules that displayed statistically significant changes that are concordant (i.e. both up or both down) between vaccine-induced transcriptomes and adjuvant-induced secretomes. Blue color signifies the biomarker candidates further assessed in vitro in the context of this study. B, For each of the three post-vaccine time points, the number of molecules displaying statistically significant changes concordant between transcriptomes and newborn/adult monocyte secretomes induced by Alum and the TLR agonists MPLA and R848. Overall, this number of concordant molecules is higher for TLRA-induced secretomes than Alum-induced secretomes; higher for adult than newborn secretomes; and higher at the 24 h time point than the later time points, suggesting concordance with our in vitro platform. Assuming the 12,893 unique genes of the Affymetrix Human Genome U133A microarray platform used in GSE18323 as the background set of commonly measurable genes/proteins in both proteomic and transcriptomic studies, we found significant number of concordant genes between GSE18323 at 24 h with the adult secretome conditions for Alum (p = 4.0e-4, Kendall's tau beta test), MPLA (p = 1.8e-7, Kendall's tau beta test) and R848 (p = 2.9e-4, Kendall's tau beta) and none for the newborn data sets at 24 h. Overall, the alignment of the transcriptomic-proteomic data sets was most significant for the adult MPLA conditions.