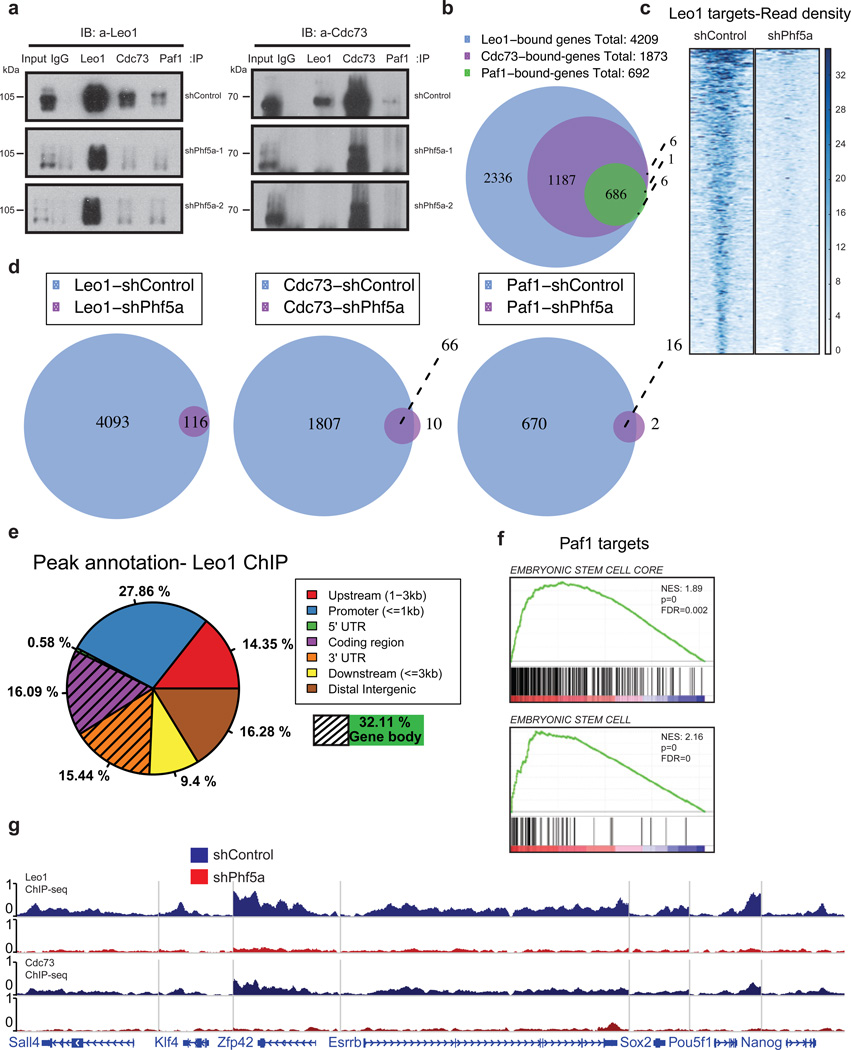
Figure 4. Phf5a controls interactions among Paf1C subunits and its silencing abrogates Paf1C recruitment on pluripotency genes in ESCs.
(a) Western blot analysis of Paf1C subunit immunoprecipitations in 293T cells following knockdown with shControl or shPhf5a, respectively, showing loss of interactions between different Paf1C members upon Phf5a depletion. Two different shRNA hairpins are shown. Left Panel: Blot for Leo1; Right Panel: blot for Cdc73; IP: immunoprecipitation. IB: immunoblot. (see Supplementary Figure 7). (b) Venn diagram showing number of genes bound by individual Paf1C subunits in ESCs using ChIP-sequencing with antibodies against endogenous Leo1, Cdc73 and Paf1 proteins. (c) Heatmap representations of normalized read density for Leo1 binding in ESCs following shControl or shPhf5a silencing, respectively. (d) Venn diagrams showing the numbers of genes bound by Leo1, Cdc73 and Paf1 in ESCs in the presence or absence of Phf5a, respectively. (e) Binding profiles for genomic distribution of Leo1 peaks (upstream, promoter, coding region, 5'UTR, 3'UTR, downstream and intergenic) in ESCs, showing preferential (32%) binding within gene bodies. (f) Gene set enrichment analysis (GSEA) enrichment plots showing significant enrichment of the top Paf1 targets for genes linked to embryonic stem cell signatures. (g) Snapshots of Leo1 and Cdc73 binding on representative pluripotency gene targets (Sall4, Klf4, Zfp42, Esrrb, Sox2, Pou5f1 and Nanog) in the presence (blue) of absence (red) of Phf5a, respectively.