Figure 2. Proteomic approach to define spatial-temporal changes in organelle proteins throughout HCMV infection.
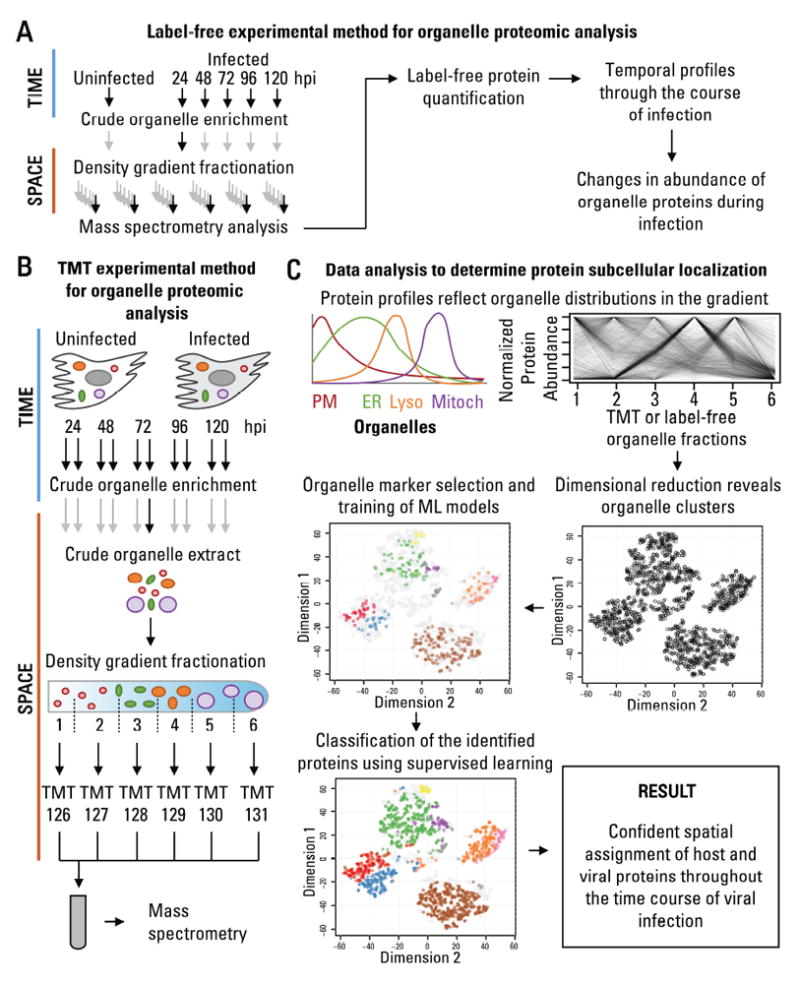
A Workflow to determine temporal changes in organelle proteins throughout infection.
B Workflow to determine changes in the subcellular localization of proteins throughout infection.
C Data analysis to determine protein subcellular localization. The relative abundance between organelle fractions collected using the methods in A or B are inspected by dimensional reduction. Manually-curated organelle markers are used to assess the quality of the organelle separation in the gradient and to determine the identity of each cluster. Organelle markers are used to train machine learning models, which then classify the remaining proteins, resulting in confident spatial assignment of proteins at each time point of viral infection.
See also fig S2.
