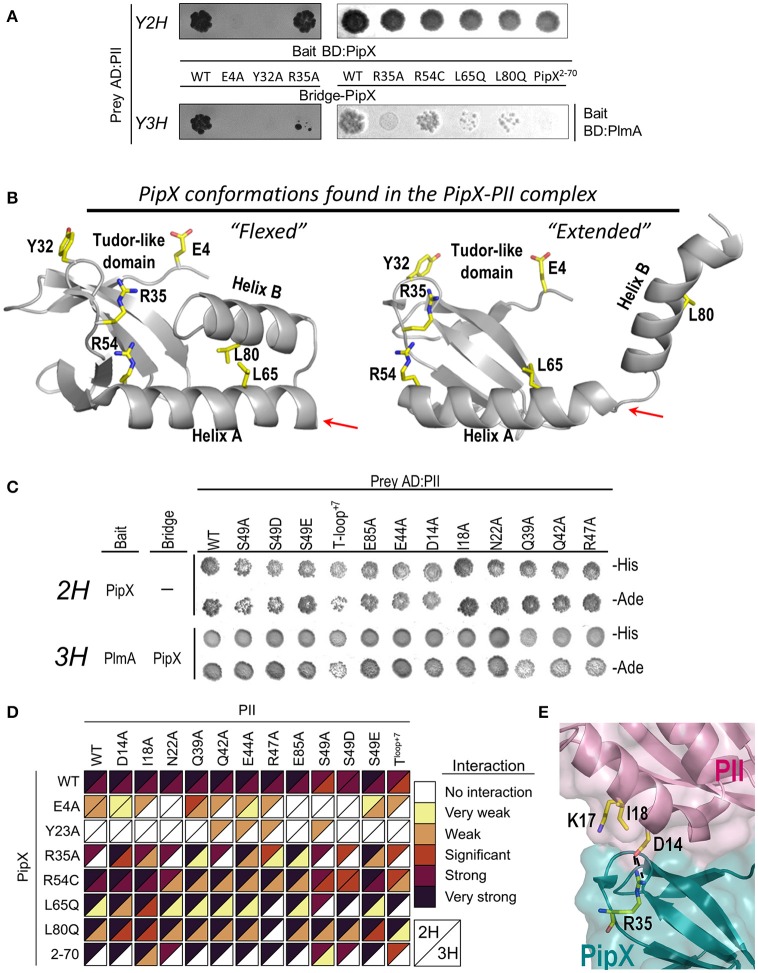
Figure 3.
Effects of PipX and PII mutations on two-hybrid and three-hybrid interaction assays. (A) Growth conferred by the indicated PipX derivatives in comparative yeast 2-hybrid (2H) and 3-hybrid (3H) assays on histidine deficient media. (B) Localization of the PipX residues mutated here on the structures of the “flexed” and “extended” conformations of this protein as observed in the PII-PipX complex of S. elongatus (taken from PDB file 2XG8) (Llácer et al., 2010). These residues are shown with their side-chains in yellow (O and N atoms red and blue, respectively) and are labeled. The right arrow marks the point of truncation in PipX2−70. (C) Effects of PII mutations in 2H and 3H assays. Pictures were taken after 4 days of incubation on histidine (-His) or adenine (-Ade) deficient media. (D) Heatmap summarizing results from 2H and 3H assays involving PII and PipX mutant derivatives. The color scale refers to the strength of the interaction signals in standard (from significant to strong interaction) and high sensitivity (from no to weak interaction) assays (see Methods). (E) Detail of PII-PipX interactions around R35 of PipX in the structure of the PII-PipX complex of S. elongatus (PDB 2XG8) to illustrate the effects of the PII mutations D14A and I18A.