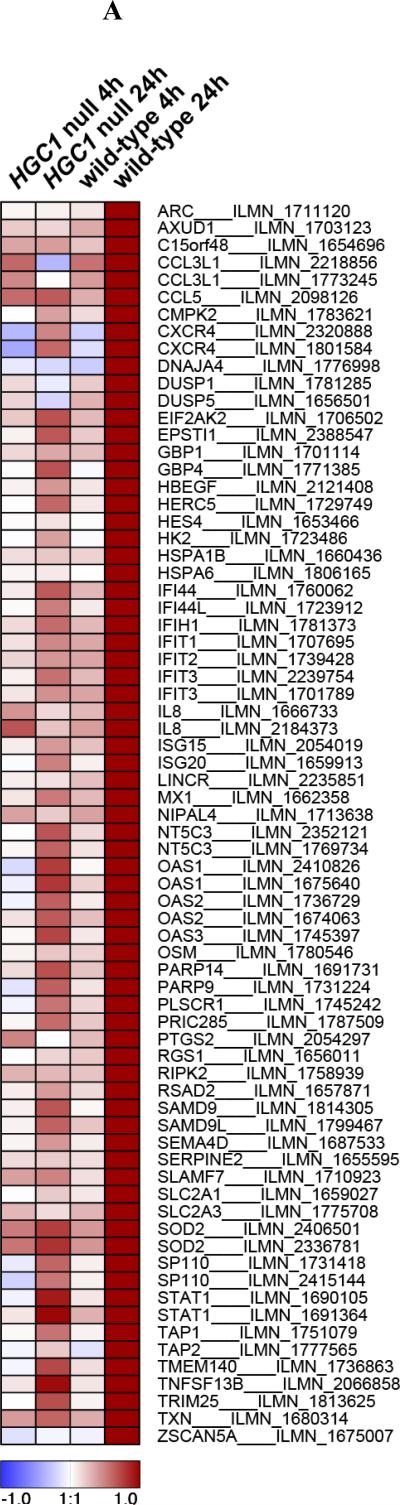
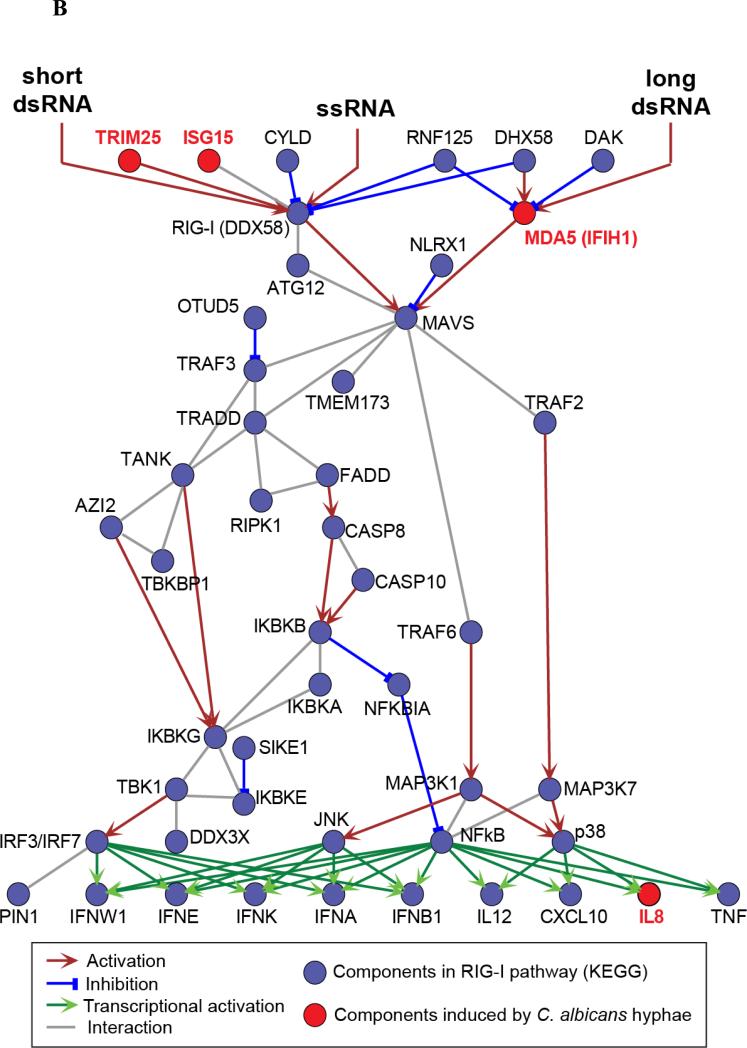
Figure 1. Transcriptional changes in macrophages stimulated with Candida albicans.
(A) The heatmap shows differential gene expression after 4h or 24h stimulation of human macrophages with yeast-locked HGC1 null Candida albicans (which are unable to form hyphae), or wild-type invasive Candida albicans (that can form hyphae), compared to expression levels in unstimulated macrophages (control). 62 genes exhibited a significant change in expression level (Benjamini-Hochberg-corrected P < 0.05 and > 2-fold change in expression) specifically after 24h stimulation with wild-type Candida, during which germination into hyphae takes place. Signal-to-noise ratio scaled to the maximum absolute deviation is shown for each probe corresponding to the 62 differentially expressed genes. (B) Candida albicans hyphae-induced genes, IFIH1, TRIM25, ISG15 and IL8 (indicated in red), are key components of the RIG-I-like receptor (RLR) signaling pathway. These genes represent both the MDA5 (IFIH1) and RIG-I (ISG15 and TRIM25) branches, as well as inflammatory cytokines that are produced by activation of the pathway (IL8). Figure based on the KEGG map of the RLR pathway [49].