Fig. 4.
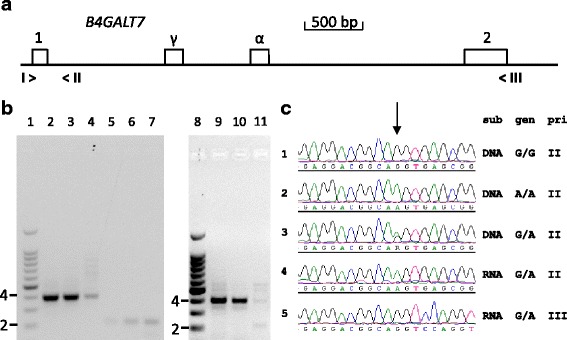
Mutation of the terminal nucleotide of exon 1 of B4GALT7 affects proper RNA splicing of intron 1. a Map of the exon 1- exon 2 region of the equine B4GALT7 gene. The positions of the exons 1 and 2 are indicated by numbered boxes; α: alternative exon; γ: cryptic exon. The position of the used PCR primers are indicated by > and < signs with Roman numerals. I>: EX1F; <II: IN1R; <III: EX2R. b cDNA fragments derived from a Friesian horse homozygous for the reference allele (lanes 2, 5 and 9), a heterozygous carrier (lanes 3. 6 and 10) and a Friesian horse dwarf (lanes 4, 7 and 11). Fibroblasts were grown from skin biopsies from the horses and RNA was isolated. cDNA was synthesized with reverse transcriptase followed by PCR with equine B4GALT7 specific primers. Lanes 1 and 8: 100 bp size standard ladder (2 = 200 bp fragment, 4 = 400 bp fragment); lanes 2-4: primers I and II; lanes 5-7: primers I and III; lanes 9-11: primers I, II and III. c Genomic DNA and RNA sequence analysis of B4GALT7 fragments from a horse homozygous for the reference allele (G/G), a dwarf (A/A) and a heterozygous carrier (G/A). The arrow indicates the position of the mutation. Sub: substrate genomic DNA (DNA) from the horses representing the three genotypes (gen) or cDNA synthesized with RNA from fibroblasts of the heterozygous horse (RNA). Pri: The PCR fragments were generated with primer I and either primer II or III as indicated. The mutant allele A is only observed in the unspliced RNA fragment (line 4) while the properly spliced product from the same heterozygous horse only shows the reference allele G (line 5)
