Figure 5.
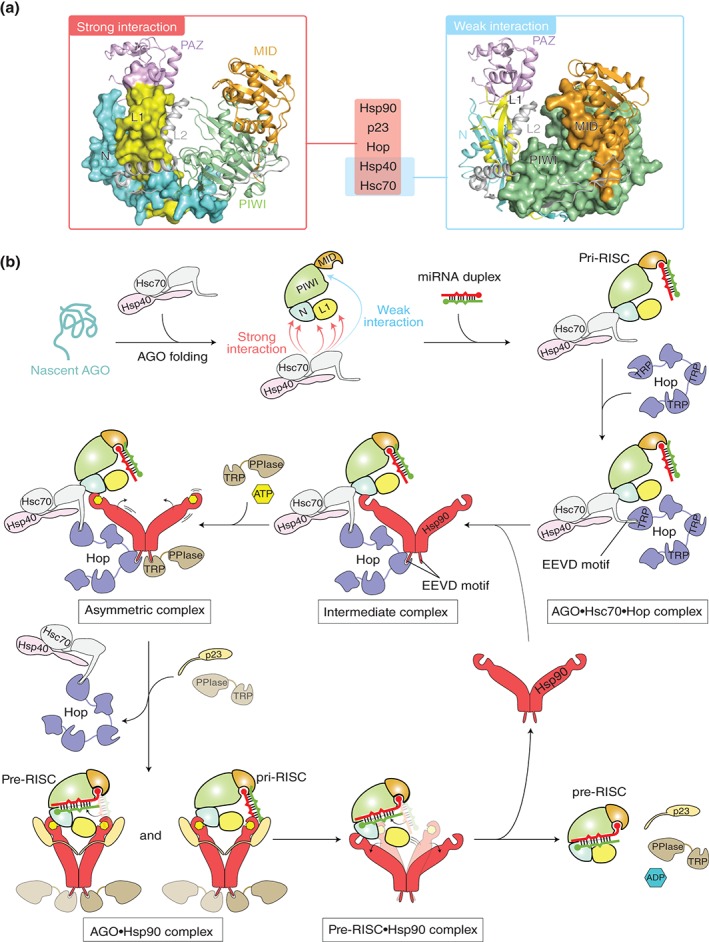
Model of chaperone‐mediated duplex loading. (a) Mapping of the chaperone‐binding sites on the structure of hAGO2 (PDB ID: 4W5N) based on the data of rat AGO256. The regions interacting strongly with Hsp90, p23, Hop, Hsp40, and Hsc70 (left) and weakly with Hsc70 and Hsp40 (right) are shown as a surface model. For clarity, the hAGO2‐bound guide RNA is not shown. (b) Duplex loading by co‐chaperone cycle of Hsp90 in humans. Nascent AGO peptide (aqua) is captured by a complex of Hsc70 (gray) and Hsp40 (pink). The PAZ domain is not shown in order to clarify the interaction between the L1 linker and chaperones. During folding of the AGO protein with the aid of the complex, Hop (slate) uses its TRP motif to recognize at the EEVD tetrapeptide of the Hsc70, which results in an AGO•Hsc70•Hop ternary complex. Hop uses another TRP motif to interact with Hsp90 (red) through the C‐terminal EEVD motif, forming an intermediate complex. Binding of ATP (yellow hexagon) to the N‐terminal domain of the Hsp90 drives the conformational changes, while a Hsp90 co‐chaperone, PPIase, containing a TRP domain (brown) binds to the remaining free EEVD motif of the Hsp90 to form an asymmetric complex. Further binding of PPIase to the C‐terminal EEVD motif of the Hsp90 results in the releases of Hop, which allows Hsp90 to capture the AGO protein. The resultant Hsp90•AGO complex in the closed conformation is stabilized by p23. The widely opened AGO protein accommodates a small RNA duplex. Probably, AGO sorts the 5′ nucleotide of the duplex using the MID domain until the bound duplex is completely accommodated into the channel (see Figure 6). ATP hydrolysis opens the structure of Hsp90 and releases ADP (blue hexagon), PPIase, and p23, along with the pre‐RISC. (Adapted from Ref 13)
