Abstract
A study on the adsorption of proteins from fetal bovine serum (FBS) on spherical dense and mesoporous silica nanoparticles with a wide range of diameters, from 70 to 900 nm, is presented. Monodisperse populations of particles with a range of diameters were obtained through modifications of the Stöber method. Extensive characterization of the particles was then performed using N2 physisorption, TEM, DLS, and ζ-potential. Following serum exposure, proteomic evaluation in concert with thermogravimetric analysis revealed the associated concentrations of each protein identified in the hard corona. Small particles adsorbed the largest amount of protein, due to their larger external surface area. Proteins with low molecular weights (<50 kDa) composed the majority of the protein corona, totaling between 60 and 80 % of the total mass of adsorbed protein. Here, the higher surface curvature of small particles favors the enrichment of smaller proteins. Porosity does not promote protein adsorption, but improves deposition of the low molecular weight protein fraction due to the size exclusion effect related to pore diameter. These results have important implications for the use of dense and porous silica nanoparticles in biomedical applications.
Keywords: protein corona, protein adsorption, silica nanoparticles, mesoporous, particle diameter, aggregation
Introduction
The integration of nanotechnology and medicine has become widespread over the last two decades, with several integrated systems currently in clinical trials.1,2 Inhibitor-loaded nanomaterials may be decorated with a multitude of targeting ligands, limiting the adverse side effects commonly observed due to the interaction of systemically-administered chemotherapeutics with normal cells. Of these materials, nano- and microparticles have received a great deal of attention due to their ease of construction and modification, biocompatibility, and uniform molecular adsorption and release kinetics.3,4 However, in order to realize the true potential of these particle-based platforms, it is important to understand the properties of the nano-bio interface.
One of the greatest challenges currently hindering the advancement of these particle-based therapies occurs when particles are initially exposed to biological fluid.5 In this environment, proteins and receptors will bind to the surface of the material, which can induce an immunogenic response,6,7 a complex process that has only recently been examined in relation to particle-based therapies.8 The proteins and biomolecules associated with particles after exposure to the biological fluid, collectively termed the ‘protein corona’, have been shown to strongly adhere to the surface.9-11 The protein corona evolves both temporally and spatially.12-14 As illustrated by Barrán-Berdón et al, “burst” adsorption, a period of rapid initial adsorption, results from proteins and biomolecules that are in high concentration and possess a strong affinity for the particle surface.12 Over time, equilibrium is reached as proteins at lower serum concentrations slowly exchange onto the particle surface. Thus, one could envision a competition for the particle surface, where each protein or molecule exhibits a unique equilibrium constant. Dynamic light scattering (DLS) studies have indicated that the protein corona is composed of both a hard inner layer, containing strongly adsorbed proteins with slow exchange rates, and a soft outer layer, containing proteins that exchange more rapidly and frequently. To date, analysis of the protein corona has primarily been conducted on the hard layer, that is, the layer of proteins remaining after the soft layer has been removed through washing. However, recently, Sakulkhu and coworkers were able to isolate the soft corona on iron oxide nanoparticles using magnetic and column separation.15 The properties of this particle-protein complex are responsible for effects on surrounding cells and tissue, rather than the native particle.
It is now widely accepted that the physicochemical properties of engineered particles greatly influence the composition of the protein corona.16,17 Size, shape, ζ-potential (“zeta” potential, related to nanoparticle surface charge in solution), and surface chemistry play a key role in the development and dynamics of the corona.18-20 The materials from which nanoparticles can be prepared, including lipids, silica, metal oxides, and various polymeric systems, each form a unique corona that depends on their chemical properties.18,21,22 In particular, porous and dense silica have received attention due to their biocompatibility and stability in a variety of biological systems. 23 , 24 By controlling synthetic parameters, properties such as particle diameter, shape, and surface composition can be easily tuned for many biomedical applications. However, while many studies have focused on silica nanoparticles (< 200 nm), larger particles (200 – 1000 nm) with the same composition and surface chemistry have been shown to exhibit different behavior in vitro and in vivo.25,26 In one example, an inverse correlation between particle size and cytotoxicity in HepG2 cells was observed, where smaller particles were found to be more cytotoxic than larger ones and 500 nm particles were found to be non-toxic.26 Similarly, a different study using mesoporous nano- and micron-sized silica particles showed that 1220 nm particles were significantly less cytotoxic than 190 and 420 nm particles in both human breast cancer cells and African green monkey kidney cells.27
Numerous reports have suggested that particle diameter is one of the key factors contributing to the formation of the protein corona. However, these reports have been limited to particles with diameters less than 200 nm.28-30 In this work, we investigate the adsorption of proteins on bare, spherical silica particles with a wide range of diameters, from 70 to 900 nm. We also compare dense and mesoporous silica particles with similar diameters. Previous work done by our group examined the relationship between surface modifications on 70 nm mesoporous silica and the identity and composition of the protein corona. These experiments will inform future studies on the relationship between protein adsorption and cytotoxicity.
Experimental Section
All materials were purchased from Sigma Aldrich and used as received unless otherwise noted. Nanoparticle morphology and size were studied by transmission electron microscopy (TEM) using a JEOL 1400 microscope operating at 80 kV. Samples were dispersed in ethanol, transferred to carbon coated copper grids, and then immediately imaged. Nitrogen gas physisorption isotherms were measured in a Micromeritics Flowsorb apparatus. Surface area calculations were carried out using the BET method, pore size distributions were calculated using to the KJS adjustment of the BJH method.31 Particle size and ζ-potential measurements were conducted by dynamic light scattering (DLS) in a Malvern Instruments Zetasizer Nano ZS. Dried materials were re-suspended in deionized water at a concentration of 5 μg/mL and measurements were performed at 25 °C. In the case of bigger particles (e.g., particle diameter of about 1 μm), for particle diameter measurement, sodium silicate 0.15% was added to the medium in order to keep a stable colloid. The mean hydrodynamic diameter was determined by cumulant analysis. The organic content on the different samples before and after protein adsorption was quantified by thermogravimetric analysis in a Mettler-Toledo TGA/SDTA851e apparatus.
Synthesis of dense 85, 250, and 500 nm silica particles
The synthesis of these particles were conducted by modifying the Stöber method.32 The following procedure describes the synthesis of 85 nm particles; Table 1 shows adjustments made for the three different particle diameters. EtOH (100 %, 20 mL) was mixed with NH4OH (13.7 M, 1.5 mL) at room temperature in a 100 mL round-bottom flask equipped with a magnetic stir bar. This mixture was stirred briefly to equilibrate, and then tetraethoxysilane (TEOS, 2 mL in 5 mL EtOH, 7 mL, 8.96 mmol) was added to the solution. This mixture was then stirred for 1 h at room temperature. Afterwards, the precipitate was isolated via centrifugation (14,800 rpm, 5 min) and resuspended in EtOH using a Branson 2510 sonicator operating at 40 kHz. This process was repeated three times in order to remove as much unreacted reagent as possible. Aggregation was avoided by suspending the nanoparticles in EtOH and sonicating for 5 – 10 mins every few days.
Table 1.
Conditions Used To Prepare 85, 250, and 500 nm Dense Silica Particles
| particle diameter | EtOH (mL) | H2O (mL) | NH4OH (M/mL) | TEOS (mL) | time (h) |
|---|---|---|---|---|---|
| 85 nm | 20.0 | -- | 13.7/1.5 | 2 + 5 mL EtOH | 1 |
| 250 nm | 46.0 | -- | 13.7/10 | 1 + 4 mL EtOH | 2 |
| 500 nm | 80.0 | 8.62 | 8.7/5.7 | 5.58 | 8 |
Synthesis of dense 900 nm silica particles
Synthesis of these particles was carried out in a two-step process. Seed particles (~300 nm) were first synthesized using a previously reported procedure, and then these particles were grown to 900 nm in situ by the slow addition of TEOS and EtOH.23 To prepare the seed particles, NH4OH (13.7 M, 10.0 mL) and EtOH (100%, 50 mL) were combined at room temperature and allowed to equilibrate. TEOS (0.5 mL in 2 mL EtOH, 0.24 mmol) was then added and the mixture was stirred for 2 h at room temperature. The seed particles now present in the flask were then grown to 900 nm by adding a solution of TEOS (7 mL in 28 mL EtOH, 33.6 mmol) slowly (~0.5 mL/min) via a separatory funnel. Following the final addition of TEOS, the particles were stirred for an additional 2 h, and then isolated by centrifugation (15000 g, 15 min) to remove most of the solvent, followed by a second centrifugation (3000 rpm, 5 min). To remove large aggregates, particles were filtered through a 5 μm filter prior to protein adsorption experiments.
Synthesis of 70 nm mesoporous silica particles
Hexadecyltrimethylammonium bromide (CTAB, 1.00 g, 2.76 mmol) was dissolved in a solution of sodium hydroxide (14 mM in H2O, 500 mL) at 80 °C and stirred rapidly. TEOS (5.0 mL, 22.4 mmol) was added over 2 minutes to the solution using a syringe pump. After 2 h, the flask was cooled in an ice bath, and the precipitate was removed by filtration and washed with water and MeOH. The solid was then dried at 100 °C overnight in an oven and calcined at 540 °C for 6 h in air.
Synthesis of 850 nm mesoporous silica particles
Mesoporous silica particles of 850 nm were synthesized according to a previously published protocol.33 EtOH (100%, 138 g), Milli-Q H2O (162 g), and NH4OH (28.95 wt%, 11.6 mL) were combined at room temperature in a round bottomed flask equipped with a magnetic stir bar. CTAB (0.280 g, 0.768 mmol) was then added and allowed to completely dissolve. After 5 min, TEOS (1.388 mL, 6.66 mmol) was added and the reaction proceeded for 2 h at room temperature. The resulting mesoporous silica particles were then isolated via centrifugation at 15000 g for 10 min to remove most of the solvent, followed by a second centrifugation at 3000 rpm for 5 min, and washed several times with EtOH and H2O. Finally, the particles were dried under vacuum at room temperature for 24 h prior to calcination. Removal of the surfactant was achieved by calcining the material at 550 °C for 6 h in air.
Protein adsorption experiments
Prior to serum exposure, particles were suspended in PBS and diluted to achieve a final concentration of 1 mg/50 μL. Subsequently, an aliquot containing 1 mg of particles was added to 10 % FBS/DMEM (1 mL). After incubation at room temperature for 1 h, the protein-adsorbed particles were isolated through centrifugation at 14800 rpm for 5 min and washed three times with PBS (1 mL) to ensure that any free or loosely bound proteins were removed from the solution.
SDS-PAGE
One-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on the proteins isolated after serum incubation. Removal of the hard corona was achieved by sonicating nanoparticles in Laemmli buffer (63 mM Tris-HCl, pH 6.8, 40 mM DTT, 0.01% (w/v) bromophenol blue, 10% glycerol, 2% (w/v) SDS). Particle suspensions were then boiled for 5-10 min in a hot water bath. Particles were then removed from the suspension through centrifugation (14,800 rpm, 5 min) and the supernatant was saved for SDS-PAGE analysis. Protein separation was then performed on a Bio-Rad Mini-PROTEAN electrophoresis system (120 V, 1.5 h). The gels were then stained for 2 h using GelCode blue stain reagent (Thermo Scientific), followed by de-staining overnight in deionized water.
Proteomics analysis
The digested peptide sample was desalted using a ZipTip C18 (P10, Millipore Corporation, Billerica, MA) according the manufacturer’s protocol, and then dried in a SpeedVac. The dried peptide samples were dissolved in 20 µl 0.1% formic acid and 2% acetonitrile, and 5 μl were loaded onto a fused silica microcapillary LC column (12 cm × 100 µm inner diameter) packed with C18 reversed-phase resin (5 μm particle size; 20 nm pore size; Magic C18AQ, Michrom Bioresources Inc.). Peptides were separated by applying a gradient of 3-60% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min for 45 min. Nanospray ESI was used to introduce peptides into a liner ion trap (LTQ)-Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanospray ionization source. Mass spectrometry data was acquired in a data-dependent acquisition mode, in which an Orbitrap survey scan from m/z 400-2000 (resolution: 30,000 FWHM at m/z 400) was paralleled by 10 LTQ MS/MS scans of the most abundant ions. After an LC-MS run was completed and spectra were obtained, the spectra were searched against the IPI Bovine protein sequence databases (V 3.85) using Proteome Discoverer software (version 1.4; Thermo Electron, San Jose, CA). The search parameters permitted a 20 ppm precursor MS tolerance and a 1.0 Da MS/MS tolerance. Oxidation of methionine (M) and carboxymethylation of cysteines (C) were allowed as variable modifications. Up to two missed tryptic cleavages of peptides were considered. The cutoffs for SEQUEST assignments were: cross-correlation (Xcorr) scores greater than 1.9, 2.5, and 3.0 for peptide charge states of +1, +2, and +3, respectively; and a delta-correlation (ΔCn) score > 0.1. Then, all .srf files for each sample were inputted into Scaffold (version Scaffold_4.0.5, Proteome Software Inc., Portland, OR) for the calculations of total spectrum counts.
Calculation of mass % of individual proteins
Following triplicate analysis of adsorbed proteins by LC-MS, the normalized spectral counts (NSpC) for each protein, which represent the percentage of each protein identified in the proteomics analysis as a function of molecular weight, were multiplied by the overall mass of adsorbed protein as determined by thermogravimetric analysis (TGA). The result of this calculation (NSpC × TGA) is the contribution of each protein to the total adsorbed mass; standard deviations were determined from these values.
Results and Discussion
To investigate the influence of particle diameter on the adsorption of proteins from serum, four batches of dense, silica spheres with diameters between 85 and 915 nm (as determined by TEM, Table 2 and Figure 1) were synthesized using modifications of the Stöber method. For simplicity, they are called 85, 250, 500, and 900 nm particles throughout this manuscript.
Table 2.
Summary of TEMa, Dynamic Light Scattering (DLS), and ζ-Potential Measurements of the Particles in Figure 1
| diameter (nm)
|
|||||
|---|---|---|---|---|---|
| sample | TEM | DLS | ζ-potential (mV) | PdI | |
| dense | a | 85 ± 9 | 137 ± 55 | −37.7 ± 5.9 | 0.123 |
| b | 251 ± 10 | 281 ± 85 | −22.6 ± 6.0 | 0.074 | |
| c | 482 ± 37 | 525 ± 201 | −37.2 ± 5.2 | 0.265 | |
| d | 914 ± 21 | 1496 ± 133 | −20.1 ± 8.6 | 0.401 | |
| porous | e | 73 ± 11 | 119 ± 60 | −12.2 ± 5.5 | 0.251 |
| f | 869 ± 45 | 754 ± 459 | −11.6 ± 4.8 | 0.371 | |
Measurements of particle diameters from TEM represent an average of 50 particles.
Figure 1.
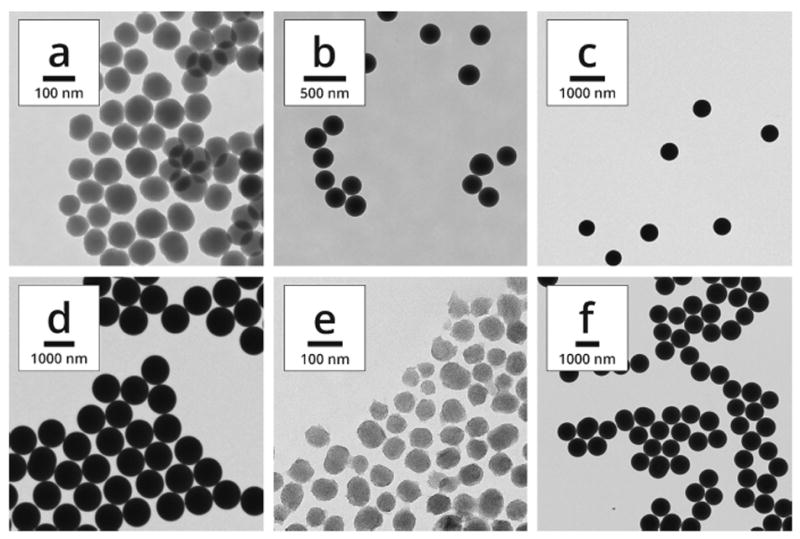
Transmission electron microscopy (TEM) images of particles with nominal diameters of (a) 85, (b) 250, (c) 500, and (d) 900 nm. Mesoporous particles of (e) 70, and (f) 850 nm are also shown. Note the different scale bars in each image.
DLS analysis was performed to analyze the ζ-potential, polydispersity index (PdI), and hydrodynamic diameter of each particle population (Table 2). Because our primary interest for this investigation was to survey the identity and composition of the protein corona as a function of particle diameter, materials with similar physicochemical properties were of great importance for both dense and mesoporous particles. For the porous materials, N2 physisorption revealed type IV isotherms for both samples, indicative of a mesoporous structure (Figure 2). The surface areas and pore diameters were consistent with expected values for mesoporous materials. Particle diameters were reasonably similar between TEM and DLS measurements with the exception of sample d; in this case, the large PdI value was an indication of the difficulty in obtaining DLS measurements for large dense particles, which can be subject to sedimentation within the optical cell in which sampling took place. Sample f showed a closer diameter between TEM and DLS measurements, although the PdI was larger, consistent with sample d. Plots of particle size distribution for all samples may be found in the Supporting Information. For consistency with the dense particles, porous particle diameters will be represented by the approximate TEM diameters (70 and 850 nm). Also of note was the difference in ζ-potential between dense and porous particles. This was most likely due to the calcination step used to remove the surfactant from samples e and f after synthesis to open the porosity. Calcination led to condensation of surface silanols and reduction of surface charge. Although ζ-potential played a part, protein adsorption appeared to be driven primarily by other factors (see below). This is consistent with the results of Mahmoudi et al., who demonstrated that other forces, i.e. protein hydrophobicity/hydrophilicity, London dispersion, protein-protein interactions, and hydrogen bonding, were the predominating factors in driving the formation of the protein corona.15
Figure 2.
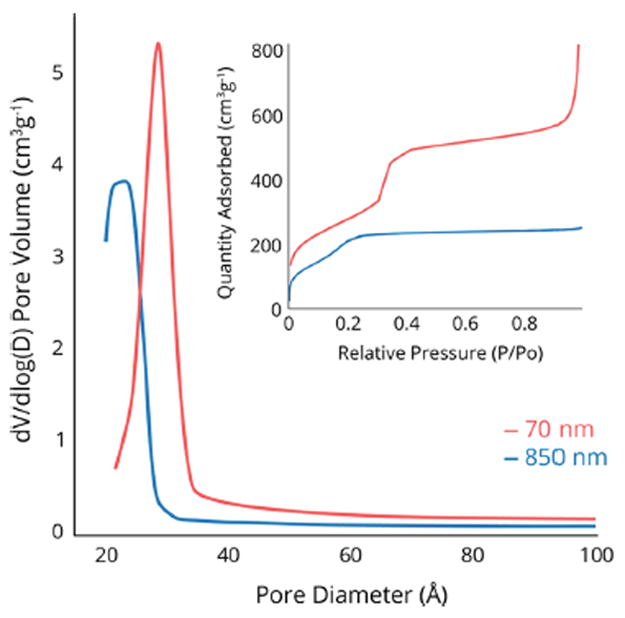
Pore size distributions of 70 nm and 850 nm mesoporous silica particles.
To evaluate the role of particle diameter on the protein corona composition and identity, particles of each diameter were incubated in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at a fixed concentration of 1 mg/mL for 1 hour at room temperature. Removal of the soft corona was achieved by isolating the particles through centrifugation, followed by repeatedly washing the particles with PBS. In order to measure the mass percentage of the remaining adsorbed proteins (i.e., the hard corona), the particle-protein complexes were dried under vacuum for at least 24 h prior to analysis. Thermogravimetric analysis (TGA) was then performed and the total amount of adsorbed protein was calculated as a function of weight loss (Table 3).
Table 3.
Summary of Protein Adsorption Data Including Normalization for Surface Area and Surface Charge
| sample | dDLS (nm) | SABET (m2g−1) | SADLS (m2g−1) | protein (mg/100 mg)a | protein per unit SADLS (mg/m2) | ζ-potential per unit SADLS (−mV/m2g−1) | |
|---|---|---|---|---|---|---|---|
| dense | a | 137 | -- | 19.9 | 10.6 | 5.33 | 1.89 |
| b | 281 | -- | 9.71 | 7.38 | 7.61 | 2.33 | |
| c | 525 | -- | 5.19 | 4.42 | 8.52 | 7.17 | |
| d | 1496 | -- | 1.82 | 4.18 | 22.9 | 11.0 | |
| porous | e | 119 | 1010 | 22.9 | 13.9 | 6.07 | 0.533 |
| f | 754 | 1012 | 3.61 | 5.22 | 14.5 | 3.21 |
From TGA.
The DLS diameter was used to calculate surface areas of dense particles because it was measured in solution. TGA analysis indicated that the smallest particles adsorbed the greatest amount of protein. This was due to differences in total surface area. For example, at 1 mg/mL, the available surface area for protein adsorption was an order of magnitude different between samples a and d. However, normalizing the total amount of adsorbed protein to the total surface area of each sample showed the opposite trend, where increasing the particle diameter greatly increased the amount of adsorbed protein. It has been hypothesized that the decreased surface curvature of larger particles favors protein binding, as proteins are able to pack together more closely on smoother surfaces.11,34 Finally, normalizing the ζ-potential to the total surface area of each sample also showed the impact of decreased surface area, with the charge increasing as particle diameter increased.
The identity and composition of the hard corona were analyzed using one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrospray liquid chromatography mass spectrometry (LC-MS/MS). SDS-PAGE analysis showed a complex mixture of proteins isolated from the particles (Figure 3). While the biomolecular fingerprint of the protein corona appeared to be similar for each particle diameter, differences in concentrations were apparent. Quantification of individual proteins was performed by combining the thermogravimetric data with the spectral counts obtained from LC-MS/MS analysis (Equations 1 & 2). In these equations, SpCk and NSpCk are the spectral counts and normalized spectral counts for an individual protein k, taken from LC-MS/MS analysis; Mwk is the molecular weight of protein k, and TGA is the weight of protein adsorbed onto the particles in μg/mg.
Figure 3.
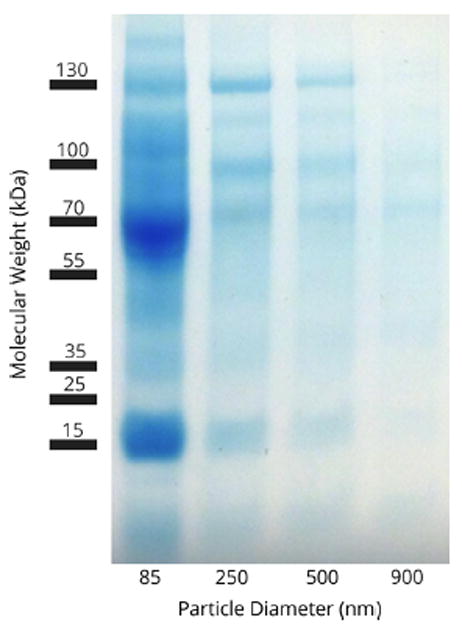
SDS-PAGE of isolated corona proteins.
| (1) |
| (2) |
For all particle diameters and types, proteins with molecular weights below 50 kDa comprised the majority of the protein corona, totaling between 60 and 80 % of the total mass of adsorbed protein (Figure 4, left panel). Interestingly, as the particle diameter increased, the mass percent of proteins with molecular weights between 50 and 100 kDa also increased, at the expense of proteins below 50 kDa. In general we observed the trend that larger particles adsorbed a greater fraction of proteins with higher molecular weights. While there appears to be an exception for the 900 nm dense particles, which adsorbed more proteins in the 50 – 100 kDa range but fewer proteins above 100 kDa, the total mass of protein adsorbed for this sample was small compared to the other particles.
Figure 4.
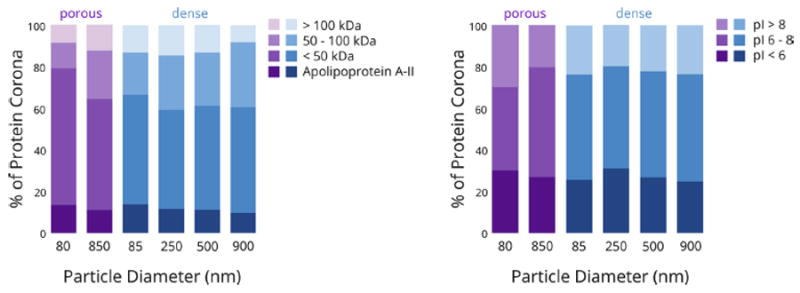
Comparison of the weight percent of the protein corona (NSpC × TGA) across dense and porous particles with different diameters, with respect to (left panel) molecular weight; (right panel) isoelectric point.
Consistent with our previous data and many other results in the literature,16,35 the amount of the proteins in the corona did not correlate with their relative abundance in the serum.36 For example, serum digestion and analysis confirmed that serum albumin was one of the most abundant proteins found in the 10% FBS used for these experiments (data not shown), but it was found in relatively low abundance on all samples that were analyzed, comprising less than 4% of the complete corona in each class of particles. Similarly, one of the most abundant serum proteins, serotransferrin, was also not identified on any particle sample. On the other hand, apolipoprotein A-II, a light protein (~11 kDa) found in high concentrations in serum, was the most abundant protein (14%) of the corona for 85 nm particles and was a major component of the corona of all particles. Here, the higher surface curvature of small nanoparticles favored the enrichment of smaller proteins;37,38 particles of 250, 500 and 900 nm were better able to accommodate heavier proteins such as apolipoprotein E (~30 kDa).
We36 and others27,39,40 have previously shown that the ζ-potential of the particles becomes less negative upon protein adsorption. Regardless of ζ-potential, none of the particles exhibited a preference for protein adsorption based on the protein’s isoelectric point (Figure 4, right panel). This is rather surprising given the negative ζ-potentials present on each type of particle. Based on electrostatic interactions, one could hypothesize that particles possessing a negative surface charge would preferentially bind positively-charged proteins. However, others have suggested that neither protein size nor charge significantly determine the protein fingerprints, confirming that electrostatic affinity alone does not constitute the major driving force regulating the silica-corona interactions.41
Protein adsorption onto porous silica particles is a more complicated process than for dense silica particles. The addition of pores along the particle surface provides openings, or void spaces, into which proteins may diffuse. Indeed, numerous investigations have reported on the immobilization of proteins within mesoporous materials.42-44 However, diffusion deep into mesoporous materials with pore diameters on the order of 4 nm (as used here) in the presence of complex protein mixtures is slow even for low molecular weight proteins, because the diameters of the folded proteins are similar to the pore diameter. Therefore, proteins are likely to gather at the pore entrances and prevent adsorption throughout the interior of large particles, so that much of the internal surface area becomes inaccessible after the first proteins are adsorbed.45,46 Some confirmation of this idea is provided by the fact that although the total surface area of the porous particles was much larger than the dense particles, the total amount of protein adsorbed (Table 3) and the trends in specific adsorption between the two groups are not very different. Also consistent with dense particles was the fact that, comparing groups of porous particles, the amount of low molecular weight proteins decreased as particle diameter increased.
On the other hand, when comparing porous particles to dense particles of the same or similar diameters, some differences become apparent. Most notable is that the fraction of low molecular weight proteins in the corona is even more enhanced when the particles are porous, regardless of particle diameter. This is particularly obvious when comparing smaller porous and dense particles; at least 80% of the proteins in the corona of the 70 nm porous silica particles had molecular weights less than 50 kDa (Figures 4 and 5). Thus in addition to the influence of surface curvature, a size exclusion effect related to the small pore diameter (~ 4 nm) also enhanced the adsorption of smaller proteins. This is consistent with our previous protein adsorption study.36
Figure 5.
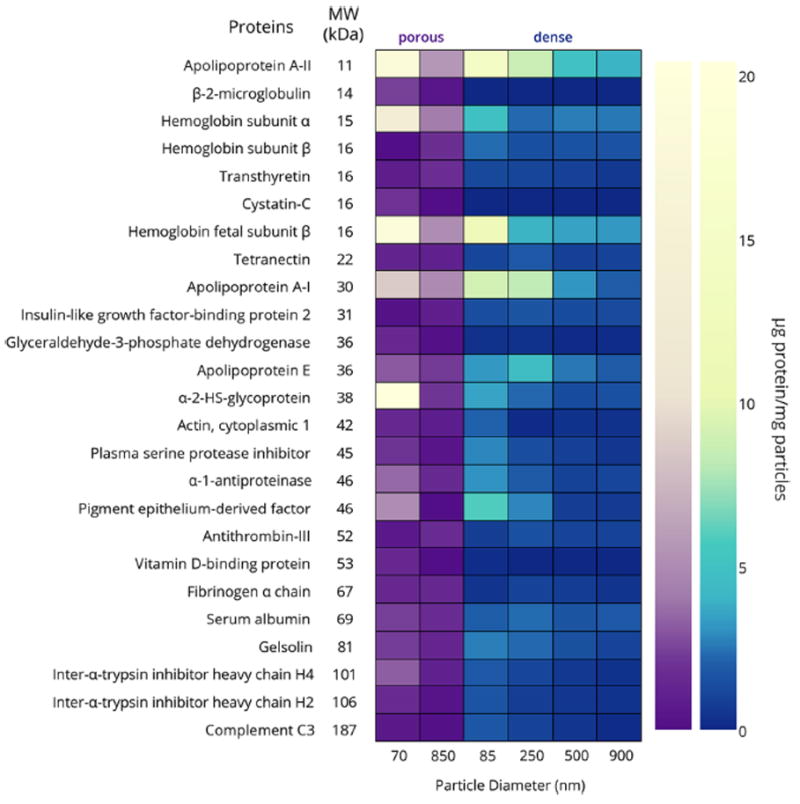
Heat map illustrating the differences in protein corona composition of porous and dense silica particles. Only proteins with concentrations greater than 1.5 μg/mg particles are included. Note that the color gradients are nonlinear to facilitate observation of proteins present in small amounts. A table of μg protein/mg particles for all identified proteins can be found in the Supporting Information.
Finally, it is interesting to note the biological function of proteins present in the corona. Consistent with other literature results, apolipoproteins comprised a majority of the protein corona on all dense and porous samples.47 Tenzer and coworkers demonstrated that these lipoproteins, which are actively involved in lipid and cholesterol transport, promote nanoparticle internalization in endothelial cells. However, in the absence of additional in vitro studies, assessing the impact of having this class of proteins in the corona is speculative. We are currently performing in vitro and in vivo studies to address issues related to particle-cell interactions.
Conclusions
Monodisperse dense and mesoporous silica particles were synthesized with a range of diameters from 70 – 900 nm in order to study the relationship between particle diameter and the formation of the protein corona. Small particles adsorbed the largest amount of protein, regardless of whether they were dense or porous. For dense particles, this was due in part to the larger surface area of the particles. Although the porous particles had nearly identical surface areas due to their large internal porosity, and therefore one might expect them to adsorb similar amounts of protein, the larger porous particles unexpectedly adsorbed significantly less protein than the smaller porous particles. This is attributed to pore blockages created by early-adsorbing proteins, which limit the surface area available to the external particle surfaces. Small dense particles adsorbed lower molecular weight proteins, due to their large surface curvature. Interestingly, porous particles of any diameter adsorbed a much greater fraction of low molecular weight proteins, due to the size exclusion effect related to pore diameter. From a practical point of view, to minimize immunogenic processes associated with protein adsorption for specific in vivo biomedical applications, silica nanoparticles with a moderate size (> 500 nm, dense or porous) are the best option. This may also be relevant in drug delivery, where particle aggregation due to protein adsorption is an important consideration.
Supplementary Material
Acknowledgments
The authors are thankful for financial support by the Spanish Ministry of Economy and Competiveness (projects SEV-2012-0267, MAT2012-39290-C02-02 and IPT-2012-0574-300000) and the University of Vermont. Research reported in this was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103449. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
A complete list of all identified proteins and the associated concentrations, as well as particle size distributions for all materials, are fully described in the Supporting Information. This information is free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Nanoparticles in Medicine: Therapeutic Applications and Developments. Clin Pharmacol Ther. 2007;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 2.Eifler A, Thaxton CS. Nanoparticle Therapeutics: FDA Approval, Clinical Trials, Regulatory Pathways, and Case Study. In: Hurst SJ, editor. Biomedical Nanotechnology. Vol. 726. Humana Press; New York: 2011. pp. 325–338. [DOI] [PubMed] [Google Scholar]
- 3.Farokhzad OC, Langer R. Nanomedicine: Developing Smarter Therapeutic and Diagnostic Modalities. Adv Drug Delivery Rev. 2006;58:1456–1459. doi: 10.1016/j.addr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding Biophysicochemical Interactions at the Nano–Bio Interface. Nat Materials. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 5.Yan Y, Gause KT, Kamphuis MMJ, Ang C-S, O’Brien-Simpson NM, Lenzo JC, Reynolds EC, Nice EC, Caruso F. Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano. 2013;7:10960–10970. doi: 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JM, Rodriguez A, Chang DT. Foreign Body Reaction to Biomaterials. Semin Immunol. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan S, Kelly PM, Mahon E, Stöckmann H, Rudd PM, Caruso F, Dawson KA, Yan Y, Monopoli MP. The “Sweet” Side of the Protein Corona: Effects of Glycosylation on Nanoparticle–Cell Interactions. ACS Nano. 2015;9:2157–2166. doi: 10.1021/nn506060q. [DOI] [PubMed] [Google Scholar]
- 8.Ritz S, Schöttler S, Kotman N, Baier G, Musyanovych A, Kuharev J, Landfester K, Schild H, Jahn O, Tenzer S, Mailänder V. Protein Corona of Nanoparticles: Distinct Proteins Regulate the Cellular Uptake. Biomacromolecules. 2015;16:1311–1321. doi: 10.1021/acs.biomac.5b00108. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Yu L, Monopoli MP, Sandin P, Mahon E, Salvati A, Dawson KA. The Biomolecular Corona is Retained during Nanoparticle Uptake and Protects the Cells from the Damage Induced by Cationic Nanoparticles until Degraded in the Lysosomes. Nanomedicine. 2013;9:1159–1168. doi: 10.1016/j.nano.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. Physical–Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. Journal of the American Chemical Society. 133:2011. 2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 11.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein—Nanoparticle Interactions: Opportunities and Challenges. Chem Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 12.Barrán-Berdón AL, Pozzi D, Caracciolo G, Capriotti AL, Caruso G, Cavaliere C, Riccioli A, Palchetti S, Laganà A. Time Evolution of Nanoparticle–Protein Corona in Human Plasma: Relevance for Targeted Drug Delivery. Langmuir. 2013;29:6485–6494. doi: 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- 13.Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Foglia P, Amenitsch H, Laganà A. Evolution of the Protein Corona of Lipid Gene Vectors as a Function of Plasma Concentration. Langmuir. 2011;27:15048–15053. doi: 10.1021/la202912f. [DOI] [PubMed] [Google Scholar]
- 14.Cedervall T, Lynch I, Lindman S, Berggàrd T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2050–2005. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein Corona Composition of Superparamagnetic Iron Oxide Nanoparticles with Various Physico-Chemical Properties and Coatings. Sci Rep. 2014:4. doi: 10.1038/srep05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rana S, Yeh Y-C, Rotello VM. Engineering the Nanoparticle–Protein Interface: Applications and Possibilities. Curr Opin Chem Biol. 2010;14:828–834. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfram J, Yang Y, Shen J, Moten A, Chen C, Shen H, Ferrari M, Zhao Y. The Nano-Plasma Interface: Implications of the Protein Corona. Colloids Surf B. 2014;124:17–24. doi: 10.1016/j.colsurfb.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DWH, Cohen Y, Emili A, Chan WCW. Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano. 20148:2439–2445. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 19.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 20.Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, McNeil SE. Interaction of Colloidal Gold Nanoparticles with Human Blood: Effects on Particle Size and Analysis of Plasma Protein Binding Profiles. Nanomedicine. 2009;5:106–117. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes VF. Hardening of the Nanoparticle–Protein Corona in Metal (Au, Ag) and Oxide (Fe3O4, CoO, and CeO2) Nanoparticles. Small. 2011;7:3479–3486. doi: 10.1002/smll.201101511. [DOI] [PubMed] [Google Scholar]
- 22.Moyano DF, Saha K, Prakash G, Yan B, Kong H, Yazdani M, Rotello VM. Fabrication of Corona-Free Nanoparticles with Tunable Hydrophobicity. ACS Nano. 2014;8:6748–6755. doi: 10.1021/nn5006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X-D, Shen Z-X, Sang T, Cheng X-B, Li M-F, Chen L-Y, Wang Z-S. Preparation of Spherical Silica Particles by Stöber Process with High Concentration of Tetra-Ethyl-Orthosilicate. J Colloid Interface Sci. 2010;341:23–29. doi: 10.1016/j.jcis.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Botella P, Abasolo I, Fernández Y, Muniesa C, Miranda S, Quesada M, Ruiz J, Schwartz S, Jr, Corma A. Surface-modified Silica Nanoparticles for Tumor-Targeted Delivery of Camptothecin and its Biological Evaluation. J Controlled Release. 2011;156:246–257. doi: 10.1016/j.jconrel.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 25.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Mesoporous Silica Nanoparticle Nanocarriers: Biofunctionality and Biocompatibility. Acc Chem Res. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Sun L, Jin M, Du Z, Liu X, Guo C, Li Y, Huang P, Sun Z. Size-Dependent Cytotoxicity of Amorphous Silica Nanoparticles in Human Hepatoma HepG2 Cells. Toxicol In Vitro. 2011;25:1343–1352. doi: 10.1016/j.tiv.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 27.He Q, Zhang Z, Gao Y, Shi J, Li Y. Intracellular Localization and Cytotoxicity of Spherical Mesoporous Silica Nano- and Microparticles. Small. 2009;5:2722–2729. doi: 10.1002/smll.200900923. [DOI] [PubMed] [Google Scholar]
- 28.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc Natl Acad Sci U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. Nanoparticle Size Is a Critical Physicochemical Determinant of the Human Blood Plasma Corona: A Comprehensive Quantitative Proteomic Analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 30.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J Am Chem Soc. 2011;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 31.Kruk M, Jaroniec M, Sayari A. Application of Large Pore MCM-41 Molecular Sieves to Improve Pore Size Analysis Using Nitrogen Adsorption Measurements. Langmuir. 1997;13:6267–6273. [Google Scholar]
- 32.Nozawa K, Gailhanou H, Raison L, Panizza P, Ushiki H, Sellier E, Delville JP, Delville MH. Smart Control of Monodisperse Stöber Particles: Effect of Reactant Addition Rate of Growth Process. Langmuir. 2005;21:1516. doi: 10.1021/la048569r. [DOI] [PubMed] [Google Scholar]
- 33.Nooney RI, Thirunavukkarasu D, Chen Y, Josephs R, Ostafin AE. Synthesis of Nanoscale Mesoporous Silica Spheres with Controlled Particle Size. Chem Mater. 2002;14:4721–4728. [Google Scholar]
- 34.Gagner JE, Lopez MD, Dordick JS, Siegel RW. Effect of Gold Nanoparticle Morphology on Adsorbed Protein Structure and Function. Biomaterials. 2011;32:7241–7252. doi: 10.1016/j.biomaterials.2011.05.091. [DOI] [PubMed] [Google Scholar]
- 35.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 36.Clemments AM, Muniesa C, Landry CC, Botella P. Effect of Surface Properties in Protein Corona Development on Mesoporous Silica Nanoparticles. RSC Adv. 2014;4:29134–29138. [Google Scholar]
- 37.Roach P, Farrar D, Perry CC. Surface Tailoring for Controlled Protein Adsorption: Effect of Topography at the Nanometer Scale and Chemistry. J Am Chem Soc. 2006;128:3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- 38.Shang W, Nuffer JH, Muñiz-Papandrea VA, Colón W, Siegel RW, Dordick JS. Cytochrome c on Silica Nanoparticles: Influence of Nanoparticle Size on Protein Structure, Stability, and Activity. Small. 2009;5:470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- 39.Caracciolo G, Callipo L, De Sanctis SC, Cavaliere C, Pozzi D, Laganà A. Surface Adsorption of Protein Corona Controls the Cell Internalization Mechanism of DC-Chol–DOPE/DNA Lipoplexes in Serum. Biochim Biophys Acta, Biomembr. 2010;1798:536–543. doi: 10.1016/j.bbamem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Lynch I, Dawson KA. Protein-Nanoparticle Interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- 41.Cárdenas B, Sánchez-Obrero G, Madueño R, Sevilla JM, Blázquez M, Pineda T. Influence of the Global Charge of the Protein on the Stability of Lysozyme–AuNP Bioconjugates. J Phys Chem C. 2014;118:22274–22283. [Google Scholar]
- 42.El-Boubbou K, Schofield DA, Landry CC. Enhanced Enzymatic Activity of OPH in Ammonium-Functionalized Mesoporous Silica: Surface Modification and Pore Effects. J Phys Chem C. 2012;116:17501–17506. [Google Scholar]
- 43.Zhou Z, Hartmann M. Progress in Enzyme Immobilization in Ordered Mesoporous Materials and Related Applications. Chem Soc Rev. 2013;42:3894–3912. doi: 10.1039/c3cs60059a. [DOI] [PubMed] [Google Scholar]
- 44.Schlipf DM, Rankin SE, Knutson BL. Pore-Size Dependent Protein Adsorption and Protection from Proteolytic Hydrolysis in Tailored Mesoporous Silica Particles. ACS Appl Mater Interfaces. 2013;5:10111–10117. doi: 10.1021/am402754h. [DOI] [PubMed] [Google Scholar]
- 45.Fan J, Yu C, Gao F, Lei J, Tian B, Wang L, Luo Q, Tu B, Zhou W, Zhao D. Cubic Mesoporous Silica with Large Controllable Entrance Sizes and Advanced Adsorption Properties. Angew Chem Int Ed. 2003;42:3146–3150. doi: 10.1002/anie.200351027. [DOI] [PubMed] [Google Scholar]
- 46.Gao F, Botella P, Corma A, Blesa J, Dong L. Monodispersed Mesoporous Silica Nanoparticles with Very Large Pores for Enhanced Adsorption and Release of DNA. J Phys Chem B. 2009;113:1796–1804. doi: 10.1021/jp807956r. [DOI] [PubMed] [Google Scholar]
- 47.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


