Abstract
CDH23 mutations have mostly been associated with prelingual severe-to-profound sensorineural hearing loss (SNHL) in either syndromic or nonsyndromic SNHL (DFNB12). Herein, we demonstrate the contribution of CDH23 mutations to postlingual nonsyndromic SNHL (NS-SNHL). We screened 32 Korean adult probands with postlingual NS-SNHL sporadically or in autosomal recessive fashion using targeted panel or whole exome sequencing. We identified four (12.5%, 4/32) potential postlingual DFNB12 families that segregated the recessive CDH23 variants, qualifying for our criteria along with rapidly progressive SNHL. Three of the four families carried one definite pathogenic CDH23 variant previously known as the prelingual DFNB12 variant in a trans configuration with rare CDH23 variants. To determine the contribution of rare CDH23 variants to the postlingual NS-SNHL, we checked the minor allele frequency (MAF) of CDH23 variants detected from our postlingual NS-SNHL cohort and prelingual NS-SNHL cohort, among the 2040 normal control chromosomes. The allele frequency of these CDH23 variants in our postlingual cohort was 12.5%, which was significantly higher than that of the 2040 control chromosomes (5.53%), confirming the contribution of these rare CDH23 variants to postlingual NS-SNHL. Furthermore, MAF of rare CDH23 variants from the postlingual NS-SNHL group was significantly higher than that from the prelingual NS-SNHL group. This study demonstrates an important contribution of CDH23 mutations to poslingual NS-SNHL and shows that the phenotypic spectrum of DFNB12 can be broadened even into the presbycusis, depending on the pathogenic potential of variants. We also propose that pathogenic potential of CDH23 variants and the clinical fate of DFNB12 may be predicted by MAF.
Introduction
Mutations of CDH23 (NM_022124) have been associated with type 1D Usher Syndrome (USH1D) and nonsyndromic hearing loss (DFNB12), in a recessively transmitted manner [1, 2]. USH1D is associated with severe manifestations, including congenital profound deafness vestibular areflexia, and visual problems due to retinitis pigmentosa. Conversely, DFNB12, is characterized by prelingual-onset nonsyndromic sensorineural hearing loss (SNHL) without the impairment of vestibular or visual functions [3].
The importance of CDH23 as a deafness gene has increasingly been recognized [4, 5]. In a Japanese study, CDH23 mutations were reported to be frequent after GJB2 and SLC26A4 in children and adults with hearing impairment [6]. Recently, CDH23 mutations have been reported in the Korean deaf population [7, 8], and genetic loads of CDH23 and its implications in the Korean pediatric population have also recently been reported [9]. Accordingly, p.P240L in CDH23 proved to exert a strong founder effect in the Korean pediatric population with severe-to-profound nonsyndromic SNHL. Although CDH23 has been well established as a deafness gene, it is challenging to delineate its function by a functional assay due to its relatively large size with 69 exons and encoded cadherin 23, which includes a protein of 3,354 amino acids with 27 extracellular cadherin (EC) domains, a single transmembrane domain, and a short cytoplasmic domain [4, 5, 10].
CDH23 related hearing loss is known to be associated with its role in the tip links of the inner ear hair cells. Tip links are extracellular filaments that is proposed to act as a gate for the mechanotransduction channel. In other words, it transduces the mechanical forces that arise from the sound waves and head movement, allowing one to hear and maintain balance [11]. The interaction between protocadherin-15 (PCDH15) and CDH23—both localized in the upper and lower parts of the tip link complex—has been reported to form tip links [12]. Accordingly, both CDH23 and PCDH15 are necessary for normal mechanotransduction, and mutations in these genes have been associated with sensory impairment [10, 13].
CDH23 related hearing loss in USH1D and DFNB12 has mostly been associated with either congenital or prelingual-onset hearing loss [5]. However, some CDH23 mutations have been reported to be associated with postlingual-onset moderate hearing loss in humans [6, 14]. Furthermore, some Cdh23 mutant alleles in mice manifested age-related hearing loss, which started as high-frequency hearing loss that eventually progressed to profound impairment with varying degrees of rapidity, as explained by the allelism and modifier gene [15–17]. Moreover, Cdh23 was also found to be susceptible to noise induced hearing loss, which is a different type of SNHL [18, 19]. However, the contribution of CDH23 to the human postlingual-onset hearing loss has not been adequately investigated. Moreover, mechanisms responsible for different phenotypes of CDH23 mutations have not been fully elucidated to date.
Herein, we adopted a genetic epidemiologic approach to objectively illustrate the importance of CDH23 in postlingual-onset SNHL and to investigate the causal relationship between genotypes and phenotypes. In brief, we estimated the carrier frequency of CDH23 mutations in postlingual adult-onset inherited hearing loss, which were segregated in either a sporadic or autosomal recessive (AR) fashion based on the ethnic-specific minor allele frequency (MAF) filtering process to investigate the contribution of CDH23 and further tried to delineate the genotypic hierarchy of CDH23 mutations that determine the fate of DFNB12.
Material and Methods
Ethical considerations
This study was approved by the Institutional Review Boards of Seoul National University Hospital (IRBY-H-0905-041-281) and Seoul National University Bundang Hospital (IRB-B-1007-105-402). Written informed consent was obtained from all study participants. In the case of minors, written informed consent was obtained from parents or guardians.
Study participants and clinical data
Patients with postlingual adult-onset SNHL, segregated in either a sporadic or AR fashion, were selected from our Korean cohorts, and published data from a previous study on pediatric cohorts were retrieved for analysis [9]. Additionally, our adult cohort fulfilled the following criteria: 1) bilateral nonsyndromic hearing loss, 2) moderate hearing loss with progressive nature, and 3) onset of hearing loss at the age of 15 or older, excluding the possibility of later development of USH1D. Also, when possible, family members were invited to participate in the study. Clinical data were obtained for this study population, including gender, age, medical history, physical examination, and audiological test results. The hearing threshold was calculated by averaging the thresholds of 0.5, 1, 2 and 4 kHz, which was classified into five categories: subtle (16–25 dB), mild (26–40 dB), moderate (41–70 dB), severe (71–95 dB), and profound (>95 dB) [20].
Molecular genetic diagnosis of postlingual SNHL
Genomic DNA was extracted from the peripheral blood samples or buccal cells, using the standard protocols (Gentra Puregene Blood Kit, Qiagen, cat. 158389; Venlo, Limburg, Netherlands). After GJB2 sequencing, we performed targeted resequencing of the known 129,200 deafness genes (TRS-129 and TRS-200), as previously described [21, 22]. TRS-129 and TRS-200 were performed by Otogenetics (http://www.otogenetics.com/) and SGI (Samsung genomic institute, http://www.samsunghospital.com/dept/main/index.do?DP_CODE=BP7), respectively. The obtained reads were aligned to the UCSC hg19 reference genome (http://genome.ucsc.edu/index.html) and variants were filtered. Further bioinformatics analyses were performed as previously described [23]. If the results of these steps were not convincing, WES was performed. Thereafter, the final candidate variants in these families were verified by Sanger sequencing and validated by ethnic-specific MAF filtering in 200 unrelated Korean control chromosomes from 100 normal hearing control subjects. Pathogenicity of the missense variants was predicted using SIFT (http://www.fruitfly.org/seq_tools/splice.html) and Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/). For an estimation of the evolutionary conservation of the amino acid sequence, we referred to the GERP++ score in the UCSC Genome Browser (http://genome.ucsc.edu/).
We considered the CDH23 variants as potentially pathogenic when they satisfied the following criteria:
CDH23 variants were not detected in the 200 normal control chromosomes (< 0.005) from our institute, which was a proposed ethnicity-specific MAF with a cut-off threshold (0.005) for autosomal recessive pathogenic variants [24, 25].
They were designated as ‘damaging or probably damaging’ by either SIFT or Polyphen-2, or a GERP++ score of higher than 3.
The residues of these variants were also conserved among several species.
A ‘probable DFNB12’ was defined as when we detected two potentially pathogenic CDH23 variants in a trans configuration that fully satisfied the above criteria. In contrast, ‘possible DFNB12’ was defined as when we were able to identify only one CDH23 variant whose pathogenic potential was previously documented.
We have deposited our whole sequencing data in our private SNUH-SNUBH sequencing database and have submitted the novel variants of CDH23, which were detected by next generation sequencing, to the Leiden Open Variation Database (LOVD) (http://databases.lovd.nl/shared/genes/CDH23).
Comparison of ethnicity-specific MAF of the rare CDH23 variants among the SNHL cohort and the normal control cohort
To further evaluate the MAF of potentially pathogenic, rare CDH23 variants in a larger size of the normal control population, a composite control cohort—which was comprised of up to 2726 normal Korean subjects (5452 alleles)—was used. The composite control cohort included 622 Korean Reference Genome (KRG) database (http://152.99.75.168/KRGDB), 700 Korean in-house exome data from the Korean National Institute of Health (KNIH), 1020 Korean control data from SGI, and 384 control individuals using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) (S1 Fig).
Phase I. First, to evaluate the contribution of potentially pathogenic, rare CDH23 variants to the Korean postlingual adult SNHL cohort, we compared the total frequency of potentially pathogenic, rare CDH23 variants with MAF of less than 0.005 between our postlingual adult SNHL cohort and the control cohort from SGI (2040 alleles). Next, we genotyped the specific CDH23 variants identified from our postlingual adult-onset SNHL cohort among the 1020 Korean control data from SGI (2040 alleles). We compared the MAF between the two groups using a Chi-square test.
Phase II. We tried to further calculate the MAF of CDH23 variants detected from our postlingual adult SNHL cohort in this study and our previously reported pediatric SNHL cohort [9] in the ethnicity-matched composite control cohort (5452 alleles). Then we compared the MAF of CDH23 variants from two cohorts among the composite control cohort using Fischer’s exact test and evaluated to see if there was any correlation between the MAF of CDH23 variants and the clinical phenotype.
Statistical analysis
Statistical analyses were performed using SPSS 18.0 (SPSS Inc., Chicago, USA). The level of statistical significance was defined as a p value of <0.05.
Results
Molecular genetic diagnosis and Clinical features of probands
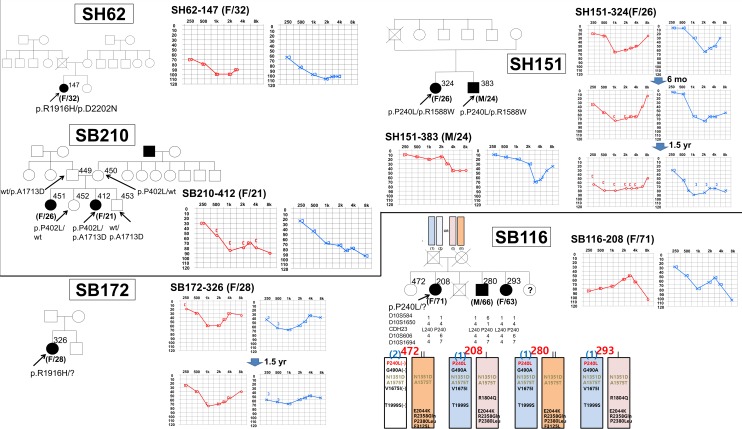
Among the 32 families of postlingual adult-onset NS-SNHL, with segregation in either a sporadic or AR fashion, we have identified four (12.5%) potential DFNB12 families: three probable DFNB12 families (SH62, SH151, and SB210) and one possible DFNB12 family (SB116). Notably, three of the four potential DFNB12 families segregated one definitely pathogenic DFNB12 variant in trans with a rare CDH23 variant with unknown pathogenicity (SH62, SH151) or with a CDH23 allele harboring a series of neighboring variants that were presumably minimally pathogenic when alone (SB116). The remaining family, SB210, co-segregated two rare CDH23 variants of unknown pathogenicity with postlingual NS-SNHL (Fig 1). The fifth family, SB172, carried only one potentially pathogenic CDH23 variant (p.R1916H), precluding any conclusive molecular diagnosis of SB172 (Fig 1).
Fig 1. Pedigrees and audiograms of subjects from 5 families possibly carrying compound heterozygous CDH23 mutations.
Audiogram: Right (red) and left (blue) ear hearing thresholds. Pedigree: Filled symbols represent hearing-impaired individuals, and clear symbols denote those with normal hearing. Black arrow indicates the proband. Possible arrangement of CDH23 variants in SB116 family based on haplotype and segregation study.
Three families (SH62, SH151, and SB210) carrying two CDH23 variants in a trans configuration manifested a rapidly progressive SNHL that started in their mid-teens to early twenties (Table 1 and Fig 1). All of the affected subjects in these families underwent cochlear implantation (CI) or were scheduled to have CI in their twenties or early thirties.
Table 1. Genotype of individuals segregating homozygous or heterozygous mutations of CDH23 identified by TRS or WES, and from the previous study by Kim et al.
[9].
| Patient | Sex/Age | Variant annotation | Location | Ref | Var | Coverage | Quality score | Found by |
|---|---|---|---|---|---|---|---|---|
| SH62-147 | F/32 | exon42:c.G5747A:p.R1916H | Chr10:73545422 | G | A | 178 | 99 | TRS200 |
| exon46:c.G6604A:p.D2202N | Chr10:73553289 | G | A | 237 | 99 | |||
| SH151-324, 383 | F/26 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 193 | 60(Qcall) | TRS129 |
| M/23 | exon37:c.C4762T:p.R1588W | Chr10:73501595 | C | T | 71 | 60(Qcall) | ||
| SB210-412 | F/21 | exon12:c.C1205T:p.P402L | Chr10:73405652 | C | T | 59 | 60(Qcall) | TRS129 |
| exon38:c.C5138A:p.A1713D | Chr10:73538016 | C | A | 21 | 60(Qcall) | |||
| SB116-208 | F/71 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 50 | 60(Qcall) | TRS129, WES |
| Not determined | ||||||||
| SH59-133 | F/3 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 238 | 99 | Kim et al. [9] |
| exon37:c.C4853A:p.T1618K | Chr10:73537445 | C | A | 29 | 99 | |||
| SH97-211 | F/1 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 102 | 60(Qcall) | Kim et al. [9] |
| exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 102 | 60(Qcall) | |||
| SH164-359 | F/1 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 218 | 99 | Kim et al. [9] |
| exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 218 | 99 | |||
| SB56-103 | F/4 | exon8:c.C719T:p.P240L | Chr10:73330641 | C | T | 238 | 60(Qcall) | Kim et al. [9] |
| exon58:c.8574delC:p.D2858EfsX8 | Chr10:73567616 | C | - | 95 | 60(Qcall) |
In contrast, SB116-208, 280, and 293 showed a progressive, moderate SNHL that started in their sixties, much later than in three aforementioned probable DFNB12 families. In this family, WES was performed on all affected family members in SB-116 to exclude other causative genes. In this family, the pathogenic p.P240L variant of CDH23 perfectly co-segregated with hearing loss phenotype, increasing the chance of CDH23 compound heterozygote being the probable candidate etiology. However, a CDH23 haplotype in trans with the p.P240L allele from one affected member (SB116-280) was different from those in the other two affected siblings (allele (II) vs (I) in Fig 1). In this family, the contribution of CDH23 was not confirmed. We checked whether any variant from either ATP2B2 or PCDH15 can contribute to hearing loss in this family in trans with the p.P240L of CDH23 as a modifier or in a digenic fashion, as previously suggested [14, 17, 26, 27]. However, none of the variants of ATPB2 or PCDH15 co-segregated with the hearing loss phenotype in this family (S1 and S2 Tables).
Lastly, one 28-year-old female (SB172-326) carrying only one potentially pathogenic CDH23 variant (p.R1916H) also complained of bilateral progressive NS-SNHL which had started 2 years ago, and had started wearing bilateral hearing aids 1.5 years ago. Her recent audiogram demonstrated progressive, moderate SNHL on both sides, in which the hearing loss pattern was similar with SH151.
Phase I: Contribution of CDH23 variants to postlingual adult-onset sporadic or arSNHL
Potentially pathogenic CDH23 variants were screened, both in our adult study and adult control cohorts. Eight alleles from six types of variants were detected: two p.P240L, one p.P402L, one p.R1588W, one p.A1713D, two p.R1916H, and one p.D2202N; the allele frequency of potentially pathogenic CDH23 variants was determined to be 12.5% (95% CI, 4.4%–20.6%)(8/64) in our postlingual adult-onset sporadic or arSNHL cohort. However, the allele frequency of all potentially pathogenic CDH23 variants in the 1020 ethnically-matched control WES data from Samsung Genome Institute (SGI) was calculated to be 5.53% (95% CI, 4.54%–6.52%) (113/2040) (S3 Table), which was significantly lower than that in our adult study cohort (p = 0.037 by Chi-square test) (S1 Fig).
Next, we focused on specific CDH23 variants, which were identified from our postlingual adult-onset SNHL cohort among the 2040 alleles from the Korean control data provided by SGI. One of the control subjects carried a p.P240L variant, and the other control subject carried a p.R1588W variant, and there were no control subjects carrying any rare CDH23 variants, such as p.R1916H, p.A1713D, p.P402L, or p.D2202N (described in Table 2 and highlighted in S3 Table). Therefore, the allele frequency of these CDH23 variants associated with postlingual adult-onset SNHL was calculated to be 0.098% (95% CI, 0%–0.23%)(2/2040) among the 1020 ethnicity-matched control subjects from SGI, which was significantly lower than that of our adult study cohort (p<0.0001 by Chi-square test) (S1 Fig).
Table 2. Ethnic-specific MAF and in silico pathogenicity prediction of CDH23 variants in our study.
| Variant | Patient | dbSNP | In-house exome from KNIH (n = 700) | KRG database (n = 622) | Genotyping (n = 384) | SGI (n = 1020) | Score in Pph2/ SIFT/ GERP | MAF in composite cohort | ExAC | 1000 Genomes |
|---|---|---|---|---|---|---|---|---|---|---|
| p.T1618K (c.C4853A) | SH59-133 (Pediatric) | No data | 0/1400 | Not detected in unknown number | 0/768 | 0/2040 | Probably Damaging/ Damaging / 5.9 | 0% (0/4208) | N/A | N/A |
| p.P240L (c.C719T) | SH59-133 SH151-324 SB116-208 (Pediatric & Adult) | rs121908354 (flagged) | 1/2040 | Possibly Damaging/ Damaging/ 5.19 | 0.05% (1/2040) | 0.00009 | 0.0002 | |||
| p.D2202N (c.G6604A) | SH62-147 (Adult) | rs121908349 (flagged) | 0/1400 | Not detected in unknown number | 0/768 | 0/2040 | Probably Damaging/ Damaging/ 5.06 | 0% (0/4208) | 0.000008 | N/A |
| p.A1713D (c.C5138A) | SB210-412 (Adult) | No data | 0/2040 | Probably Damaging/ Damaging/ 5.4 | 0% (0/2040) | N/A | N/A | |||
| p.P402L (c.C1205T) | SB210-412 (Adult) | rs373168635 (Non-flagged) | 0/2040 | Probably Damaging/ Tolerated/ 4.91 | 0% (0/2040) | 0.00003 | N/A | |||
| p.R1588W (c.C4762T) | SH151-324 (Adult) | rs137937502 | 4/1400 | 2/1244 | 1/768 | 1/2040 | Probably Damaging/ Damaging/ 3.24 | 0.15% (8/5452) | 0.0002 | 0.0008 |
| p.R1916H (c.G5747A) | SH62-147 (Adult) | rs746971522 | 3/1400 | 2/518 | 0/764 | 0/2040 | Probably Damaging/ Damaging/ 4.28 | 0.11% (5/4722) | 0.00006 | N/A |
KRG, Korean Reference Genome (http://152.99.75.168/KRGDB); ExAC, Exome Aggregation Consortium (http://exac.broadinstitute.org/); 1000 Genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/); N/A, not applicable
Phase II: Comparison of MAF between CDH23 variants detected in adult study cohort and pediatric study cohort among ethnicity-matched controls
Three adult patients (SH62-147, SH151-324, and SB210-412) carried two potentially pathogenic CDH23 variants as a compound heterozygote, whereas four pediatric patients (SH59-133, SH97-211, SH164-359, and SB56-103) carried two CDH23 variants either in homozygous or compound heterozygous state (Table 1) [9]. We focused on the pathogenicity of each CDH23 allele, which were determined by their ethnic-specific MAF in our composite cohort (adult control cohort) (S1 Fig) [24, 25].
Ethnic-specific MAF of seven variants of CDH23 in our adult and pediatric study cohorts was displayed (Table 2). MAF of a missense variant, p.T1618K, of CDH23, which was detected from a pediatric cohort in trans configuration to p.P240L in SH59-133, was 0% (0/4208), while that of p.R1588W and p.R1916H, which were in trans to p.P240L and p.D2202N from adult patients, was 0.15% (95% CI, 0.05%–0.25%) (8/5452) and 0.11% (95% CI, 0.02%–0.2%) (5/4722), respectively, in our composite adult cohort (Table 2). The ethnic-specific MAF of p.T1618K detected from the pediatric SNHL population was significantly lower than that of p.R1588W and p.R1916H detected from the postlingual-onset adult SNHL population (p = 0.03, and 0.05 respectively by Fischer’s exact test). None of the control individuals carried p.D2202N, p.A1703D, and p.P402L.
Discussion
Some CDH23 mutations in humans have previously been reported to be associated with adult-onset postlingual progressive SNHL, in both Caucasians and Japanese [6, 14]. However, systematic documentation of the contribution of CDH23 mutations to this late-onset postlingual progressive SNHL was not a main concern in these two reports. Instead, the role of a modifier gene, ATP2B2, was elucidated to account for the phenotypic differences among siblings [14]. Miyagawa et al. (2012) did not rigorously investigate the causal relationship between the CDH23 genotype and its phenotype [6]. In contrast, our current study, which employs a genetic epidemiologic approach, clearly demonstrated that an alteration of the CDH23 gene contributes to adult-onset postlingual progressive NS-SNHL. The rare CDH23 alleles that satisfy our criteria for a potential pathogenicity were more frequently detected in Korean adult-onset postlingual progressive SNHL than in normal hearing controls, with statistical significance. In fact, this result is not surprising since the association of age-related progressive SNHL and some Cdh23 alleles, such as Cdh23ahl and Cdh23erl, has already been documented in mouse models with certain genetic backgrounds [17, 18, 28]. The replacement of a single nucleotide (A to G) in a Cdh23 gene on progressive SNHL had been shown to prevent age-related SNHL phenotype [6, 17, 29, 30].
Based on our result, we could think that the major form of CDH23-related SNHL might be adult-onset progressive SNHL, rather than prelingual-onset severe-to-profound SNHL (DFNB12), at least in Koreans or East Asians. In fact, CDH23 mutations accounted for 9.4% (3/32)—or possibly up to 12.5% (4/32)—of postlingual adult-onset SNHL, while the genetic load of CDH23 mutations was 3.1% (4/128) in our pediatric cohort with prelingual-onset severe-to-profound SNHL [9]. A statistical analysis was performed to compare the frequency of CDH23 mutation between the adult and pediatric hearing loss cohorts; a difference was shown with marginal significance (p = 0.051 by Fischer’s exact test). From this observation, we can assert that CDH23 variants might contribute more to adult-onset progressive SNHL than to prelingual-onset severe-to-profound SNHL. However, we need to be cautious to draw a firm conclusion from these results due to the relatively small number of subjects in this study.
One important finding from our study is that the p.P240L allele of CDH23, which turned out to be the founder allele among the prelingual DFNB12 Korean subjects, was revisited in our adult-onset postlingual SNHL cohort. Two (SH151 and SB116) of the five families with co-segregating adult-onset progressive SNHL with at least one potentially pathogenic CDH23 variant turned out to have the p.P240L allele. A contribution of the p.P240L allele to our adult-onset postlingual SNHL was confirmed by a higher frequency of this allele (2/64) in our adult SNHL cohort than in normal controls (1/2040) (p<0.0001 by Fischer’s exact test). The p.P240L homozygotes were previously reported to cause prelingual severe-to-profound SNHL in a majority of cases [6, 9, 31]. According to our study, as well as previous Japanese studies, audiological phenotypes of CDH23 compound heterozygotes that carry one p.P240L allele seem to be highly variable [6, 31]. Indeed, SH151-324 carrying p.P240L/p.R1588W manifested progressive SNHL, which started from mid to high frequencies at the age of 20. In contrast, SB116-208 showed progressive SNHL that became noticeable after the age of 60 years in the present study. Given this, it can be postulated that auditory phenotypes of these families depend on the pathogenic potential or residual CDH23 protein dosage from the trans CDH23 allele to p.P240L. In our previous study with a pediatric pre-lingual SNHL population, the trans allele of p.P240L was p.P240L itself, p.D2858EfsX8, which was a truncation mutation, and p.T1618K, respectively. It is easily conceivable that p.D2858EfsX8 leads to a serious deleterious effect on proteins. The MAF of this missense variant, p.T1618K, was extremely low as indicated by zero detection among 4208 control alleles, implying a strong pathogenic potential [32]. Among the two postlingual SNHL families (SH151 and SB116) carrying p.P240L, the trans allele of p.P240L of CDH23 from SH151-324 was p.R1588W; however, none of the potential polymorphisms of CDH23 in trans with p.P240L was compatible with the segregation of SNHL in SB116. The pathogenic potential of p.R1588W of CDH23 was previously disputed due to the presence of normal hearing from one homozygous carrier of p.R1588W [6]. However, it is likely that p.R1588W, presumably with a mild pathogenicity, can exert its pathogenic effect leading to progressive postlingual SNHL when in trans with a strongly pathogenic p.P240L.
The role of CDH23 mutations in SB116 is enigmatic. The CDH23 haplotype of trans allele to p.P240L was not shared by all the siblings with SNHL in this family. This may suggest that mutations in CDH23 do not account for SNHL in SB116 and that the detection of p.P240L was fortuitous. However, some of the frequent neighboring single nucleotide polymorphisms of CDH23 were indeed shared en bloc by all three siblings with SNHL (Fig 1). The collective effects of these SNPs in CDH23 might exert a pathogenic effect, albeit not severe, in a trans configuration with p.P240L, leading to progressive SNHL prominent after the 60’s. A perfect co-segregation of p.P240L of CDH23 with the SNHL phenotype in SB116 supported this hypothesis. Significantly late onset age of SNHL in SB116 might also increase the possibility of a relationship between CDH23 and presbycusis in an older population.
Differential auditory phenotype, depending on the combination of the two Cdh23 alleles in mice, showed a striking resemblance to our observation. Homozygous mice carrying the functionally null mutations of Cdh23, such as Cdh23v or Cdh23v-ngt, manifest congenital profound SNHL and a severe vestibular phenotype [33, 34]. In contrast, homozygosity with respect to Cdh23753A (hypomorphic Ahl allele) leads to increased susceptibility to age-related SNHL, thereby showing severe hearing loss by 9–12 months of age, under certain genetic backgrounds [18]. Interestingly, the compound heterozygous mice with one null allele of Cdh23v-ngt and one hypomorphic allele (Cdh23ahl) under C57BL/6J showed an intermediate phenotype: hearing impairment that started from the age of 4 months which increased in severity in an age-dependent manner [35]. Taken together, the combination of two Cdh23 alleles with a different pathogenic potential decided the fate of auditory phenotype in mice.
From this perspective, we looked into the human CDH23 variants that we detected from our present study, as well as from our previous study by Kim et al. (2015) [9]. First, we made a comparison of the ethnic-specific MAF between the missense variants detected from our adult SNHL cohort and the missense variants from our pediatric SNHL cohort. We observed significantly higher MAF of p.R1588W and p.R1916H from the postlingual-onset adult SNHL population than that of p.T1618K, which was in trans with p.P240L [9] in a pre-lingual profound SNHL. The most likely explanation would be that the difference in the pathogenic toxicity of the two alleles (p.R1588W vs p.T1618K) in trans with the known pathogenic allele, p.P240L, decided the fate of auditory phenotype in two subjects. The milder pathogenic potential of p.R1588W as compared with p.T1618K could be indirectly supported by normal hearing from the p.R1588W homozygous carrier [6] and also by the higher Korean MAF [32]. Another two missense variants, p.D2202N (rs121908349 flagged) and p.A1713D, in postlingual adult-onset SNHL, which were detected in trans with p.R1916H and p.P402L, respectively, may exert just as strong pathogenic potential as p.P240L, especially considering the GERP score of more than 5, in silico prediction results and extremely low MAF (Table 2). Additionally, the pathogenic potential of p.D2202N was previously described [1]. In contrast, p.R1916H and p.P402L (rs373168635) may also serve as milder pathogenic alleles, considering their relatively low GERP score of <5, MAF, and in silico prediction results (Table 2). From this explanation, it is possible to suggest MAF as a predictor of the clinical fate of DFNB12.
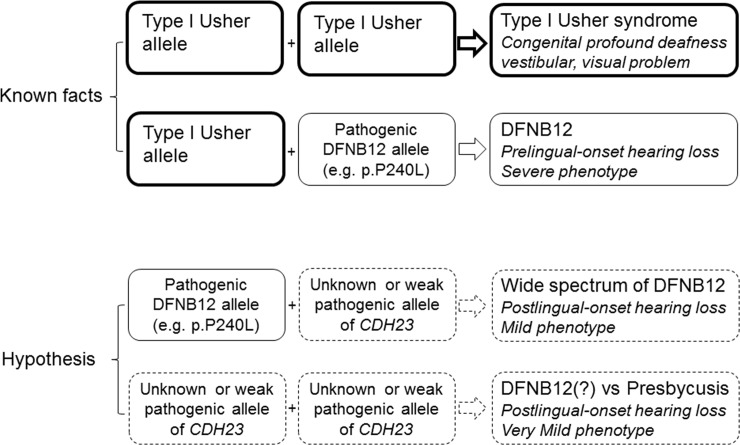
In a previous genotype-phenotype correlation study, it was suggested that the phenotypic consequence of the compound heterozygosity for a DFNB12 allele in trans configuration with a predicted USH1D allele of CDH23 was determined, mostly by the DFNB12 allele [5]. In this present study, we tried to extend this hypothesis to include the phenotypic consequence of the compound heterozygosity of a known pathogenic DFNB12 allele in trans configuration with CDH23 allele with an unknown pathogenicity, or to even include the compound heterozygosity of two CDH23 alleles with an unknown pathogenicity (Fig 2).
Fig 2. Suggestion of Genotypic Hierarchy of CDH23 mutations deciding the fate of USH1D and DFNB12.
Conclusions
This study shows an important contribution of CDH23 variants to postlingual NS-SNHL. It also broadens the phenotypic spectrum of DFNB12 to include even presbycusis depending on the allelic combinations of CDH23. We also propose a potential role of Korean MAF of rare CDH23 variants as a predictor of pathogenic potential and clinical fate of DFNB12 in postlingual NS-SNHL. These patients carrying such CDH23 variants need to be counseled about cochlear implantation, based on the nature of rapid progression to severe-to-profound SNHL irrespective of age at diagnosis. Furthermore, since it is possible to correct age-related hearing loss in mice by repairing a single mutation in the Cdh23 allele, it may be feasible to treat CDH23-related hearing loss in humans with gene therapy in the near future [29].
Supporting Information
Phase I: Comparison of carrier frequency of potentially pathogenic CDH23 alleles between our postlingual adult SNHL cohort and the control cohort. Phase II: Comparison of MAF between CDH23 variants detected in postlingual adult SNHL. cohort and prelingual pediatric SNHL cohort among the composite control cohort.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files. Also, we have submitted the novel variants of CDH23 that we detected by next generation sequencing to the Leiden Open Variation Database (LOVD) (http://databases.lovd.nl/shared/genes/CDH23) (submission numbers 0000081436 and 0000081437).
Funding Statement
This work was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (www.mohw.go.kr/) (HI14C1867 and HI15C1632 to BYC); and also by the Korea National Institute of Health intramural research grant 4861-307-210-13 (2012-NG61004-00 to SKK); and Korea Biobank Project (http://www.nih.go.kr/NIH_NEW/main.jsp) (4851-307, KBP-2014-035 to SKK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. American journal of human genetics. 2001;68(1):26–37. 10.1086/316954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nature genetics. 2001;27(1):108–12. 10.1038/83667 . [DOI] [PubMed] [Google Scholar]
- 3.Friedman TB, Schultz JM, Ahmed ZM, Tsilou ET, Brewer CC. Usher syndrome: hearing loss with vision loss. Advances in oto-rhino-laryngology. 2011;70:56–65. 10.1159/000322473 . [DOI] [PubMed] [Google Scholar]
- 4.Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. American journal of human genetics. 2002;71(2):262–75. 10.1086/341558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, et al. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. Journal of medical genetics. 2011;48(11):767–75. 10.1136/jmedgenet-2011-100262 . [DOI] [PubMed] [Google Scholar]
- 6.Miyagawa M, Nishio SY, Usami S. Prevalence and clinical features of hearing loss patients with CDH23 mutations: a large cohort study. PloS one. 2012;7(8):e40366 10.1371/journal.pone.0040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo HM, Park HJ, Park MH, Kim BY, Shin JW, Yoo WG, et al. Identification of CDH23 mutations in Korean families with hearing loss by whole-exome sequencing. BMC medical genetics. 2014;15:46 10.1186/1471-2350-15-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JH, Kim NK, Kim AR, Rhee J, Oh SH, Koo JW, et al. Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet journal of rare diseases. 2014;9:167 10.1186/s13023-014-0167-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SY, Kim AR, Kim NK, Kim MY, Jeon EH, Kim BJ, et al. Strong founder effect of p.P240L in CDH23 in Koreans and its significant contribution to severe-to-profound nonsyndromic hearing loss in a Korean pediatric population. Journal of translational medicine. 2015;13:263 10.1186/s12967-015-0624-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, et al. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428(6986):950–5. 10.1038/nature02483 . [DOI] [PubMed] [Google Scholar]
- 11.Pickles JO, Comis SD, Osborne MP. Cross-links between stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hearing research. 1984;15(2):103–12. . [DOI] [PubMed] [Google Scholar]
- 12.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449(7158):87–91. 10.1038/nature06091 . [DOI] [PubMed] [Google Scholar]
- 13.Alagramam KN, Goodyear RJ, Geng R, Furness DN, van Aken AF, Marcotti W, et al. Mutations in protocadherin 15 and cadherin 23 affect tip links and mechanotransduction in mammalian sensory hair cells. PloS one. 2011;6(4):e19183 10.1371/journal.pone.0019183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz JM, Yang Y, Caride AJ, Filoteo AG, Penheiter AR, Lagziel A, et al. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N Engl J Med. 2005;352(15):1557–64. 10.1056/NEJMoa043899 . [DOI] [PubMed] [Google Scholar]
- 15.Hequembourg S, Liberman MC. Spiral ligament pathology: a major aspect of age-related cochlear degeneration in C57BL/6 mice. Journal of the Association for Research in Otolaryngology: JARO. 2001;2(2):118–29. 10.1007/s101620010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane KL, Longo-Guess CM, Gagnon LH, Ding D, Salvi RJ, Johnson KR. Genetic background effects on age-related hearing loss associated with Cdh23 variants in mice. Hearing research. 2012;283(1–2):80–8. 10.1016/j.heares.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh RK, Friedman RA. Genetics of hearing loss: Allelism and modifier genes produce a phenotypic continuum. The anatomical record Part A, Discoveries in molecular, cellular, and evolutionary biology. 2006;288(4):370–81. 10.1002/ar.a.20297 . [DOI] [PubMed] [Google Scholar]
- 18.Noben-Trauth K, Zheng QY, Johnson KR. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nature genetics. 2003;35(1):21–3. 10.1038/ng1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holme RH, Steel KP. Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. Journal of the Association for Research in Otolaryngology: JARO. 2004;5(1):66–79. 10.1007/s10162-003-4021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark JG. Uses and abuses of hearing loss classification. ASHA. 1981;23(7):493–500. . [PubMed] [Google Scholar]
- 21.Oh SK, Choi SY, Yu SH, Lee KY, Hong JH, Hur SW, et al. Evaluation of the pathogenicity of GJB3 and GJB6 variants associated with nonsyndromic hearing loss. Biochimica et biophysica acta. 2013;1832(1):285–91. 10.1016/j.bbadis.2012.05.009 . [DOI] [PubMed] [Google Scholar]
- 22.Kim BJ, Kim AR, Han KH, Rah YC, Hyun J, Ra BS, et al. Distinct vestibular phenotypes in DFNA9 families with COCH variants. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies. 2016. 10.1007/s00405-015-3885-1 . [DOI] [PubMed] [Google Scholar]
- 23.Chang MY, Kim AR, Kim NK, Lee C, Lee KY, Jeon WS, et al. Identification and Clinical Implications of Novel MYO15A Mutations in a Non-consanguineous Korean Family by Targeted Exome Sequencing. Mol Cells. 2015;38(9):781–8. 10.14348/molcells.2015.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, Gurrola J 2nd, Simpson A, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. American journal of human genetics. 2014;95(4):445–53. 10.1016/j.ajhg.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moteki H, Azaiez H, Booth KT, Shearer AE, Sloan CM, Kolbe DL, et al. Comprehensive genetic testing with ethnic-specific filtering by allele frequency in a Japanese hearing-loss population. Clinical genetics. 2015. 10.1111/cge.12677 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, et al. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14(1):103–11. 10.1093/hmg/ddi010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed ZM, Kjellstrom S, Haywood-Watson RJ, Bush RA, Hampton LL, Battey JF, et al. Double homozygous waltzer and Ames waltzer mice provide no evidence of retinal degeneration. Mol Vis. 2008;14:2227–36. [PMC free article] [PubMed] [Google Scholar]
- 28.Han F, Yu H, Tian C, Chen HE, Benedict-Alderfer C, Zheng Y, et al. A new mouse mutant of the Cdh23 gene with early-onset hearing loss facilitates evaluation of otoprotection drugs. Pharmacogenomics J. 2012;12(1):30–44. 10.1038/tpj.2010.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mianne J, Chessum L, Kumar S, Aguilar C, Codner G, Hutchison M, et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome medicine. 2016;8(1):16 10.1186/s13073-016-0273-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohmen J, Kang EY, Li X, Joo JW, Hormozdiari F, Zheng QY, et al. Genome-wide association study for age-related hearing loss (AHL) in the mouse: a meta-analysis. Journal of the Association for Research in Otolaryngology: JARO. 2014;15(3):335–52. 10.1007/s10162-014-0443-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizutari K, Mutai H, Namba K, Miyanaga Y, Nakano A, Arimoto Y, et al. High prevalence of CDH23 mutations in patients with congenital high-frequency sporadic or recessively inherited hearing loss. Orphanet journal of rare diseases. 2015;10:60 10.1186/s13023-015-0276-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–69. 10.1038/nrg2344 . [DOI] [PubMed] [Google Scholar]
- 33.Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nature genetics. 2001;27(1):103–7. 10.1038/83660 . [DOI] [PubMed] [Google Scholar]
- 34.Wada T, Wakabayashi Y, Takahashi S, Ushiki T, Kikkawa Y, Yonekawa H, et al. A point mutation in a cadherin gene, Cdh23, causes deafness in a novel mutant, Waltzer mouse niigata. Biochem Biophys Res Commun. 2001;283(1):113–7. 10.1006/bbrc.2001.4724 . [DOI] [PubMed] [Google Scholar]
- 35.Miyasaka Y, Suzuki S, Ohshiba Y, Watanabe K, Sagara Y, Yasuda SP, et al. Compound heterozygosity of the functionally null Cdh23(v-ngt) and hypomorphic Cdh23(ahl) alleles leads to early-onset progressive hearing loss in mice. Exp Anim. 2013;62(4):333–46. 10.1538/expanim.62.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phase I: Comparison of carrier frequency of potentially pathogenic CDH23 alleles between our postlingual adult SNHL cohort and the control cohort. Phase II: Comparison of MAF between CDH23 variants detected in postlingual adult SNHL. cohort and prelingual pediatric SNHL cohort among the composite control cohort.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Also, we have submitted the novel variants of CDH23 that we detected by next generation sequencing to the Leiden Open Variation Database (LOVD) (http://databases.lovd.nl/shared/genes/CDH23) (submission numbers 0000081436 and 0000081437).