Abstract
To identify metastasis-related proteins in nasopharyngeal carcinoma (NPC), iTRAQ-tagging combined with 2D LC-MS/MS analysis was performed to identify the differentially expressed proteins (DEPs) in high metastatic NPC 5-8F cells and non-metastatic NPC 6-10B cells, and qRT-PCR and Western blotting were used to confirm DEPs. As a result, 101 DEPs were identified by proteomics, and 12 DEPs were selectively validated. We further detected expression of three DEPs (RAN, SQSTM1 and TRIM29) in a cohort of NPC tissue specimens to assess their value as NPC metastatic biomarkers, and found that combination of RAN, SQSTM1 and TRIM29 could discriminate metastatic NPC from non-metastatic NPC with a sensitivity of 88% and a specificity of 91%. TRIM29 and RAN expression level were closely correlated with lymph node and distant metastasis and clinical stage (P <0.05) in NPC patients. Finally, a combination of loss-of-function and gain-of-function approaches was performed to determine the effects of TRIM29 on NPC cell proliferation, migration, invasion and metastasis. The results showed that TRIM29 knockdown significantly attenuated while TRIM29 overexpression promoted NPC cell in vitro proliferation, migration and invasion and in vivo metastasis. The present data first time show that SQSTM1, RAN and TRIM29 are novel potential biomarkers for predicting NPC metastasis, demonstrate that TRIM29 is a metastasis-promoted protein of NPC.
Keywords: nasopharyngeal carcinoma, metastasis, iTRAQ, TRIM29, SQSTM1, RAN
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is one of the most common malignant tumors in southern China and Southeast Asia. Early metastasis is one of distinctive characteristics of NPC [1]. Although NPC is sensitive to radiotherapy, significant rates of relapse and distant metastasis still occur after therapy, which is a major cause for NPC lethality [2]. The five-year survival rate following combined treatment with radiotherapy and adjuvant cisplatin chemotherapy is 50-60% and the rates of five-year cumulative local relapse and distant metastasis are 20-30 and 20-25%, respectively [3]. However, the molecular mechanism of NPC metastasis is not yet well-defined. To develop better diagnosis and treatment approaches, it is important to identify proteins related to NPC metastasis and understand the molecular basis of NPC metastasis.
Both 5-8F and 6-10B NPC cell lines derived from human NPC low-differentiated squamous cancer cells (SUNE1), which have the same genetic background but not the same metastatic potentials. It has been confirmed that 5-8F possesses high metastatic potential, but 6-10B possesses non- metastatic potential. Therefore, the two cell lines are often used to study the molecular events of NPC metastasis [4, 5].
Proteomics has become a new subject for investigating protein expression and activities. It can comprehensively, dynamically, and quantitatively observe the change of protein expression in the process of tumor development and metastasis [6]. Isobaric tags for relative and absolute quantitation (iTRAQ) in combination with two-dimensional liquid chromatography tandem MS (2D-LC-MS/MS) analysis is emerging as one of the more powerful policy for proteomics methodologies in the search for tumor biomarkers [7–10]. This new approach possesses many advantages compared with the traditional proteomic technologies, such as high throughput, high repeatability, high sensitivity, and high accuracy [7]. Although this approach has been widely applied to screen proteins correlated with NPC [11–15], there are no reports about screening NPC metastasis-associated proteins by using this approach.
The tripartite motif (TRIM) family is composed of multidomain ubiquitin E3 ligases, and characterized by the presence of the N-terminal tripartite motif (RING, B-boxes, and coiled coil). Currently, more than 70 members of TRIM family have been identified in humans. These proteins participate in cell growth and development regulation, and have been implicated in several human diseases such as HIV infection and leukemia [16, 17]. TRIM29 is a member of the tripartite motif (TRIM) family [18]. Several studies have shown that TRIM29 plays an important role in the various types of cancers [19–27]. However, to our best knowledge, the functions of TRIM29 in NPC have never been characterized.
To search biomarkers for early detection of NPC metastasis, in this study iTRAQ tagging followed by 2D LC-MS/MS was performed to identify DEPs between high metastatic 5-8F and non-metastatic 6-10B NPC cells, some DEPs identified by proteomics were selectively validated, the value of three DEPs (RAN, SQSTM and TRIM29) for predicting NPC metastasis was assessed, and the effects of TRIM29 on NPC cell proliferation, migration and invasion were determined.
RESULTS
Identification of differentially expressed proteins (DEPs) between 5-8F and 6-10B cell using iTRAQ labeling and 2D-LC-MS/MS
A total of 862 non-redundant proteins were repeatedly identified by iTRAQ labeling and 2D-LC-MS/MS analyses in NPC cells, 79.37% of which were identified with ≥2 peptides matches. The detailed information including information of peptide sequences, protein quantification date, and average iTRAQ ratio for these identified proteins is shown in supplementary Table S-3.
To identify the DEPs related to NPC metastasis, protein expressional profiles between the two cell lines were compared. The proteins which met the following criteria were confidently considered as DEPs: (1) proteins were repeatedly identified by the twice experiments; (2) proteins were identified based on ≥2 peptides; (3) proteins showed an average ratio-fold change ≥1.2 or ≤ 0.8 and the same change trend in the twice experiments between the two cells (t-test; P < 0.05), as a result, 101 proteins were found to be differentially expressed. The names of the 101 proteins are shown in Supplementary Table S-4. MS/MS spectra used for the identification and quantitation of RAN, SQSTM1, and TRIM29 are shown in Figure 1A.
Figure 1. MS/MS spectra of RAN, SQSTM1, and TRIM29 and Expressional changes of TRIM29, RAN, and SQSTM1 in NPC cells.
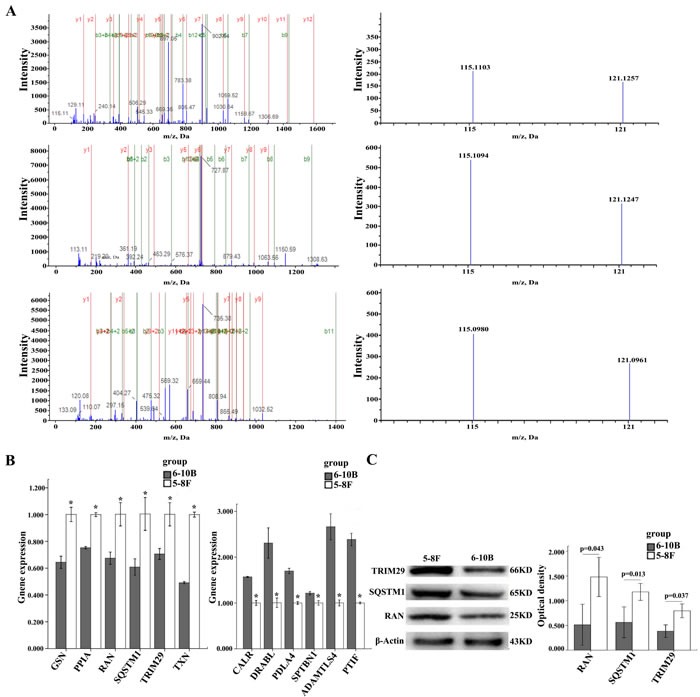
A. (above) RAN (SNYNFEKPFLWLAR M/Z 697.0439 Z = 3), the sequence SNYNFEKPFLWLAR allows the identification of RAN(right); the released iTRAQ reporter ions provide the relative quantitation of RAN from the two cells evaluated(left). (middle) TRIM29 (SALFAGNEWR M/Z 727.8883 Z = 2), the sequence SALFAGNEWR allows the identification of TRIM29(left); the released iTRAQ reporter ions provide the relative quantitation of TRIM29 from the two cells evaluated(right). (below) SQSTM1 (VAALFPALRPGGFQAHYR M/Z 569.5746 Z = 4), the sequence VAALFPALRPGGFQAHYR allows the identification of SQSTM1(left); the released iTRAQ reporter ions provide the relative quantitation of SQSTM1 from the two cells evaluated(right). 5-8F, labeled with iTRAQ reagent 115; 6-10B, labeled with iTRAQ reagents 121. B. The mRNA expressional levels of twelve genes (GPX1, NPM1, CALR, PTGES3, VCP, AIMP1, PPIA, GSN, SQSTM1, TXN, TRIM29, RAN) which were consistent with MS analysis. histogram shows the gene relative expression levels .(left) Expressions of six genes increased in 5-8F compared to 6-10B; *P < 0.05, by Independent sample T test. (right) Expressions of other six genes decreased in 5-8F compared to 6-10B. *P < 0.05, by Independent sample T test; Bars = means ± SD. C. A representative result of Western blotting showing the expressions of RAN, SQSTM1, and TRIM29 in the 5-8F and 6-10B (left); histogram shows the expression levels of the three proteins in two kinds of cells as determined by densitometric analysis (right). Actin is used as the internal loading control. Bars = means ± SD; *P < 0.05; by Independent sample T test.
Validation of DEPs indentified by quantitative proteomics
Through Uniprot and Pubmed search, 12 different proteins (GSN, PPIA, RAN, SQSTM1, TRIM29, TXN, CALR, DIABLO, PDIA4, SPTBN1, ADAMTSL4, PTRF) identified by MS analysis were chosen in 101 DEPs for verification. QRT-PCR was performed to detect their expression in 5-8F and 6-10B cells. As shown in Figure 1B, the expression changes of their mRNAs are consistent with the findings in MS analysis. In addition, we chose three proteins (RAN, SQSTM1, TRIM29) by Uniprot and Pubmed search for further validation using Western blot. The result showed that the expression of these three proteins was significantly higher in 5-8F cells than that in 6-10B cells (Figure 1C), which are also consistent with the results of MS analysis.
Value of RAN, SQSTM1 and TRIM29 as biomarkers for predicting NPC metastasis
To investigate whether the three proteins (RAN, SQSTM1, TRIM29) serve as biomarkers for predicting NPC metastasis, immunohistochemistry was performed to detect the expressional levels of the three proteins in an cohort of formalin-fixed and paraffin-embedded archival tissue specimens including metastatic and non-metastatic NPCs. As shown in Figure 2A and Table 1, the expression levels of all the three proteins were increased in the metastatic NPCs compared with non-metastatic NPCs, which also supports our findings in the NPC cells.
Figure 2. Expressional changes of TRIM29, RAN, and SQSTM1 in NPC tissues and their efficacy in discriminating metastatic NPC from non-metastatic NPC.
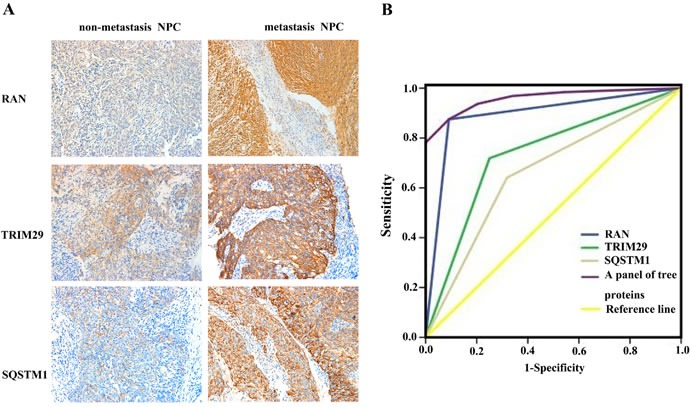
A. A representative result of immunohistochemistry shows the expression of RAN, SQSTM1, and TRIM29 in the metastatic NPC and non-metastatic NPC. Original magnification: ×200. B. Receiver operating characteristic (ROC) curves of RAN, SQSTM1, TRIM29 in discriminating metastatic NPC from non-metastatic NPC.
Table 1. Expression of RAN, SQSTM1, and TRIM29 expression in 108 nasopharygeal carcinomas.
| Non-metastatic NPC | Metastatic NPC | |
|---|---|---|
|
SQSTM1 High(5-8) |
14 | 41 |
| Low(0-4) | 30 | 23 |
|
RAN High(5-8) |
22 | 56 |
| Low(0-4) | 22 | 8 |
|
TRIM29 High(5-8) |
11 | 44 |
| Low(0-4) | 33 | 20 |
P<0.01, by Mann-Whitney U test, Non-metastatic NPC vs. Metastatic NPC.
The ability of these three proteins in distinguishing metastatic NPC from non-metastatic NPC was analyzed by determining the ROC curves individually and as a panel. The area under the curve (AUC) of these proteins is listed in the Table 2 together with their individual and collective values of merit. When individual protein serves as a biomarker, their sensitivity and specificity are 64-88% and 68-91% in discriminating metastatic NPC from non-metastatic NPC, respectively. As a panel, these proteins achieved a sensitivity of 88% and a specificity of 91% in discriminating metastatic NPC from non-metastatic NPC (Figure 2B; Table 2).
Table 2. Receiver operating characteristics of the three proteins from IHC scores, for distinguishing metastatic NPC from non-metastatic NPC (individually and as a panel).
| Proteins | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|
| RAN | 0.88 | 0.91 | 0.93 | 0.83 | 0.89 |
| TRIM29 | 0.72 | 0.75 | 0.81 | 0.65 | 0.73 |
| SQSTM1 | 0.64 | 0.68 | 0.75 | 0.57 | 0.66 |
| A panel of three proteins | 0.88 | 0.91 | 0.93 | 0.83 | 0.96 |
AUC: Area under the curve; NPV: Negative predict value; PPV: Positive predict value.
The relationship between the expression levels of RAN, SQSTM1 and TRIM29 and clinicopathological characteristics of NPC patients
The relationship between the expression levels of RAN, SQSTM1 and TRIM29 and clinicopathological characteristics in patients with NPC are summarized in Tables 3∼5. We observed that RAN and TRIM29 expression level were closely correlated with lymph node (P = 0.000; P = 0.000) and distant metastasis (P = 0.002; P = 0.000), and clinical stage (P = 0.036; P = 0.041); SQSTM1 expression level was closely correlated with lymph node metastasis (P = 0.002), but not correlated with distant metastasis (P = 0.073) and clinical stage (P = 0.733) in NPC patients. We did not find any significant association of RAN, SQSTM1 and TRIM29 expression with age, sex, and primary lesion size in patients with NPC. The results indicated that high expression of the three proteins enhanced NPC metastasis.
Table 3. Association between RAN expression and clinicopathological characteristics in 108 nasopharyngeal carcinomas.
| Variable | Expression level | |||
|---|---|---|---|---|
| n | Low(0-4) | High(5-8) | P | |
| Gender | ||||
| Male | 72 | 20 | 52 | 1.000 |
| Female | 36 | 10 | 26 | |
| Age(y) | ||||
| <50 | 66 | 18 | 48 | 0.883 |
| ≥50 | 42 | 12 | 30 | |
| Primary tumor(T) stage | ||||
| T1-2 | 30 | 11 | 19 | 0.201 |
| T3-4 | 78 | 19 | 59 | |
| lymph node(N) metastasis | ||||
| N0 | 45 | 22 | 23 | 0.000# |
| N1-3 | 63 | 8 | 55 | |
| Distant metastasis(M) | ||||
| M0 | 77 | 28 | 49 | 0.002# |
| M1 | 31 | 2 | 29 | |
| Clinical TNM stage | ||||
| I-II | 19 | 9 | 10 | 0.036* |
| III-IV | 89 | 21 | 68 | |
P<0.01 by χ2 test, N0 vs.N1-3; M0 vs.M1.
P<0.05 by χ2 test, TNM stage I-II vs. III-IV.
Table 5. Association between TRIM29 expression and clinicopathological characteristics in 108 nasopharyngeal carcinomas.
| Variable | Expression level | |||
|---|---|---|---|---|
| n | Low(0-4) | High(5-8) | P | |
| Gender | ||||
| Male | 72 | 33 | 39 | 0.683 |
| Female | 36 | 18 | 18 | |
| Age(y) | ||||
| <50 | 66 | 33 | 33 | 0.469 |
| ≥50 | 42 | 18 | 24 | |
| Primary tumor(T) stage | ||||
| T1-2 | 30 | 15 | 15 | 0.720 |
| T3-4 | 78 | 36 | 42 | |
| lymph node(N) metastasis | ||||
| N0 | 45 | 34 | 11 | 0.000# |
| N1-3 | 63 | 17 | 46 | |
| Distant metastasis(M) | ||||
| M0 | 77 | 45 | 32 | 0.000# |
| M1 | 31 | 6 | 25 | |
| Clinical TNM stage | ||||
| I-II | 19 | 13 | 6 | 0.041* |
| III-IV | 89 | 38 | 51 | |
P<0.01 by χ2 test, N0 vs.N1-3; M0 vs.M1.
P<0.05 by χ2 test, TNM stage I-II vs. III-IV.
Table 4. Association between SQSTM1 expression and clinicopathological characteristics in 108 nasopharyngeal carcinomas.
| Variable | Expression level | |||
|---|---|---|---|---|
| n | Low(0-4) | High(5-8) | P | |
| Gender | ||||
| Male | 72 | 35 | 37 | 0.892 |
| Female | 36 | 18 | 18 | |
| Age(y) | ||||
| <50 | 66 | 33 | 33 | 0.809 |
| ≥50 | 42 | 20 | 22 | |
| Primary tumor(T) stage | ||||
| T1-2 | 30 | 14 | 16 | 0.848 |
| T3-4 | 78 | 38 | 40 | |
| lymph node(N) metastasis | ||||
| N0 | 45 | 30 | 15 | 0.002# |
| N1-3 | 63 | 23 | 40 | |
| Distant metastasis(M) | ||||
| M0 | 77 | 42 | 35 | 0.073 |
| M1 | 31 | 11 | 20 | |
| Clinical TNM stage | ||||
| I-II | 19 | 10 | 9 | 0.733 |
| III-IV | 89 | 43 | 46 | |
P<0.01 by χ2 test, N0 vs.N1-3.
The effects of TRIM29 on NPC cell proliferation, and in vitro migration and invasion
The significant increase of TRIM29 expression in the NPC cell and tissues with high metastatic potentials hinted us to investigate its role in NPC cell metastasis both in vitro and in vivo. We generated 5-8F NPC cell lines with knockdown of TRIM29 and 6-10B NPC cell lines with overexpression of TRIM29 (Figure 3A). MTT assay showed that proliferation of TRIM29 knockdown 5-8F cells significantly decreased compared with control cells (Figure 3B). Conversely, proliferation of TRIM29 overexpression 6-10B cells significantly decreased compared with control cells (Figure 3B). Wound healing and transwell invasion assay showed that TRIM29 knockdown inhibited migration and invasion of 5-8F cells (Figure 3C and 3D), whereas TRIM29 overexpression promoted migration and invasion of 6-10B cells(Figure 3C and 3D). The results suggested that TRIM29 plays a key role in NPC cell migration and invasion in vitro.
Figure 3. The effects of TRIM29 on NPC cell proliferation, and in vitro migration and invasion.
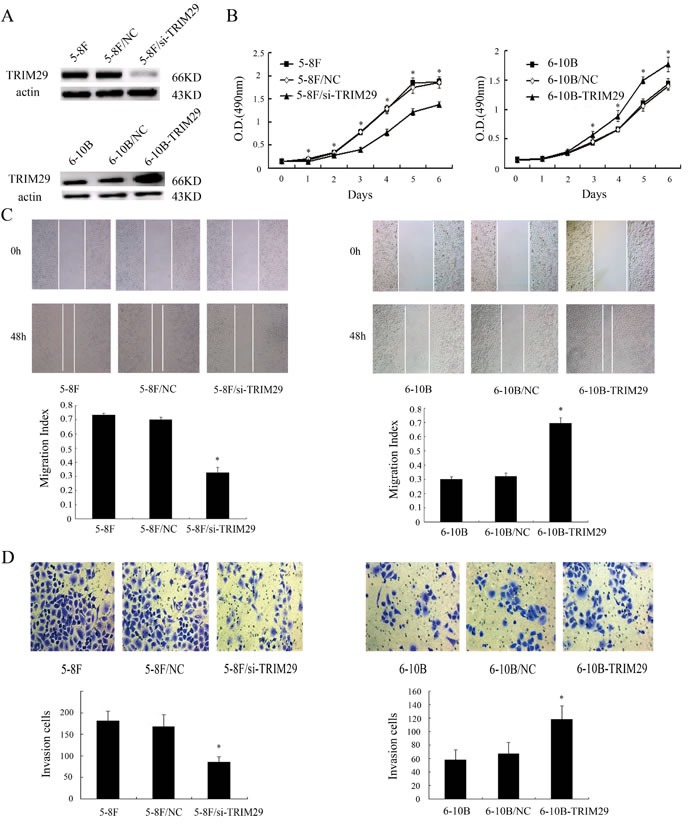
A. (above) A representative result of Western blotting shows the levels of TRIM29 expression in the untransfected (5-8F), empty vector (5-8F/NC) and TRIM29 siRNA plasmid transfected 5-8F cells (5-8F/si-TRIM29). (below) A representative result of Western blotting shows the levels of TRIM29 expression in the untransfected(6-10B), empty vector (6-10B/NC) and TRIM29 expressing plasmid transfected 6-10B cells (6-10B-TRIM29). Actin is used as the internal loading control. B. (left) MTT shows knockdown of TRIM29 in 5-8F cells inhabits cell proliferation. (right) MTT shows overexpression of TRIM29 in 6-10B cells promotes cell proliferation. Data were presented as mean±SD(*P < 0.05; by one-way ANOVA). C. (left) A representative result of scratch wound-healing assay shows knockdown of TRIM29 in 5-8F cells inhibits cell migration in vitro. (right) A representative result of scratch wound-healing assay shows overexpression of TRIM29 in 6-10B cells promotes cell migration in vitro. D. (left) A representative result of transwell assay shows knockdown of TRIM29 in 5-8F cells inhibits cell invasion in vitro. (right) A representative result of transwell assay shows overexpression of TRIM29 in 6-10B cells promotes cell invasion in vitro.
The effect of TRIM29 on NPC cell metastasis in vivo
To explore the role of TRIM29 in NPC metastasis in vivo, we tested the effect of TRIM29 in a xenograft metastasis model in which TRIM29 knockdown 5-8F cells, TRIM29 overexpression 6-10B cells, and their corresponding control cells were used to generate pulmonary metastases in nude mice. As shown in Figure 4, pulmonary metastases generated by TRIM29 knockdown 5-8F cells were significantly less than those generated by control 5-8F cells, whereas pulmonary metastases generated by TRIM29 overexpression 6-10B cells were significantly more than those generated by control 6-10B cells. The results suggested that TRIM29 plays a key role in NPC cell metastasis in vivo.
Figure 4. The effect of TRIM29 on NPC cell metastasis In Vivo.
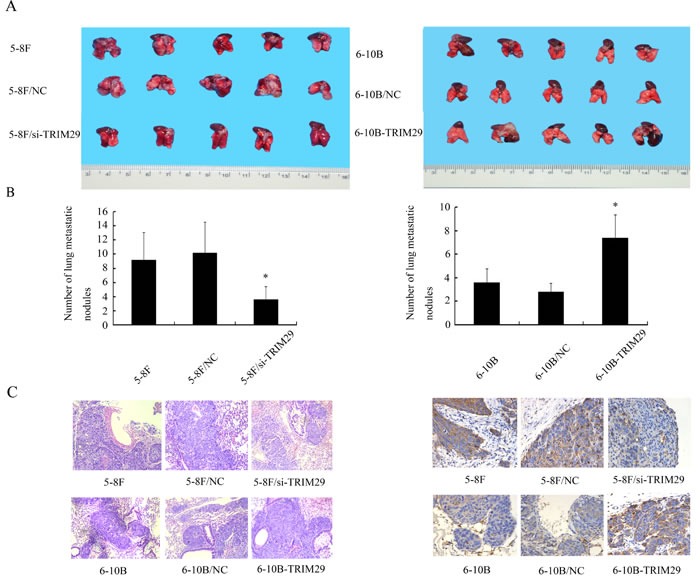
A. In vivo metastasis assays of NPC cells with TRIM29 expression changes. TRIM29 knockdown 5-8F cells, TRIM29 overexpression 6-10B cells, and their corresponding empty vector-transfected cells,and5-8F cells, 6-10B cells were injected into the tail vein of nude mice, and after 6 weeks the reprentative photography of lung was shown.(left) Knockdown of TRIM29 inhibits metastasis of NPC cells in vivo. (right) Overexpression of TRIM29 promotes metastasis of NPC cells in vivo. B. Histogram of average numbers of lung metastases per mouse. (left) Numbers of lung metastatic nodules in nude mice injected with 5-8F/si-TRIM29 cells is less than 5-8F and 5-8F/NC. (right) Numbers of lung metastatic nodules in nude mice injected with 6-10B-TRIM29 cells is less than injected with 6-10B cells and 6-10B/NC cells. C. (left) A representative result of HE shows tumor metastatic condition in 5-8F, 5-8F/NC and 5-8F/si-TRIM29. Original magnification, ×100. (right) A representative result of HE shows tumor metastatic condition in 6-10B, 6-10B/NC and 6-10B-TRIM29. Original magnification, ×100. D. (left) A representative result of immunohistochemistry shows the expression of TRIM29 in 5-8F, 5-8F/NC and 5-8F/si-TRIM29. Original magnification, ×400. (right) A representative result of immunohistochemistry shows the expression of TRIM29 in 6-10B, 6-10B/NC and 6-10B-TRIM29. Original magnification, ×400.
Gene-ontology, KEGG pathways analysis and PPI of the differential proteins
For an overview of the differential proteins in NPC metastasis, functional analysis including cellular component, molecular functions, biological process and pathways were performed using MAS 3.0 tools. The cellular component category showed that differential proteins are mainly involved in cytoplasm, nucleus, cytosol and mitochondrion. In the molecular function category, DEPs are mainly associated with binding, catalysis, molecular transduction, transport, and molecular structure. The biological process category indicated that the differential proteins are mainly related to signal transduction, transport, cell redox homeostasis, oxidation reduction, RNA splicing and regulation of transcription(Supplementary table 5). An pathway enrichment analysis was conducted by KEGG for 101 differential proteins. By this means, we identified 11 pathways significantly enriched with association signals at the p < 0.01 level. These pathways encompassed metabolism, infection, human diseases, ribosome and regulation of actin cytoskeleton. They may play a key role in the process of nasopharyngeal carcinoma metastasis(Supplementary Figure 1). Supplementary Figure 2 shows the PPI generated by STRING 9.5. Differential proteins tightly linked to each other, they interacted and jointly promote the process of nasopharyngeal carcinoma.
DISCUSSION
Identification of proteins related to NPC metastasis is important in discovery of biomarkers for early warning NPC metastasis. In this study, iTRAQ labeling combined with 2D-LC-MS/MS was used to identify DEPs in the NPC cells with different metastatic potentials. As a result, 101 DEPs were identified, and 12 DEPs were selectively validated. Next, we evaluated the ability of three DEPs (RAN, SQSTM1, TRIM29) for predicting NPC metastasis, and found that panel of these three proteins could greatly distinguish metastatic NPC from non-metastatic NPC with high sensitivity and specificity, suggesting that the three proteins are potential biomarkers for early warning NPC metastasis.
RAN, a GTP-binding nuclear protein, member of small GTPase superfamily, is involved in several important cellular processes, the best known being nucleocytoplasmic transportation [30], Which regulates many cancer-related signaling pathways such as PI3K/Akt/mTORC1 and MEK/ERK, and plays an important role in cancer cell survival and progression [31, 32]. It is overexpressed in various cancers with prognostic significance [33, 34] and its overexpression is correlated with increased aggressiveness of the cancer cells in vitro and in vivo [35, 36]. Fan H et al. also found that RAN expression increases in colorectal cancer tissues compared with normal colorectal mucosa, and was positively associated with invasion depth, lymph node and distant metastasis, and differentiated degree of the tumors [37]. Our iTRAQ results showed that RAN expression levels increased in NPC tissues with metastasis compared with ones without metastasis, and were positively associated with lymph node and distant metastasis, and clinical stage in NPC patients, supporting that RAN is a metastasis-promoted protein.
SQSTM1, also known as EBI3-associated protein of 60 kDa (EBIAP) or ubiquitin-binding protein p62, regulates a lot of biological processes such as apoptosis, cell differentiation, autophagy, protein localization, and protein ubiquitination [38–42]. It can activate many cancer-related signaling pathways such as NF-kB pathway and MAPK pathway, mTOR pathway, and promote the proliferation and spread of cancer cells [43–49]. SQSTM1 has been identified as a component of inclusion bodies found in various human diseases including HCC and malignant glioma [50, 51]. Furthermore, the loss of SQSTM1 suppressed Ras-induced lung adenocarcinoma [45]. Moreover, dysregulation of NF-kB signal in autophagy- incompetent cells was due, at least in part, to accumulation of SQSTM1, which subsequently enhanced tumorigenesis [52]. Those lines of evidence suggest the involvement of aberrant signals related to p62 accumulation in tumorigenesis. Our results also showed that high SQSTM1 expression positively correlated with NPC metastasis. To our best knowledge, it is first report on SQSTM1′s role in NPC.
Table 6. The absorbance values of 5-8F cells after transfecion (490 nm).
| cells | Day | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
| 5-8F | 0.145±0.011 | 0.190±0.028 | 0.337±0.014 | 0.772±0.049 | 1.277±0.093 | 1.842±0.119 | 1.876±0.065 |
| 5-8F/NC | 0.150±0.009 | 0.206±0.028 | 0.340±0.035 | 0.790±0.054 | 1.287±0.080 | 1.749±0.129 | 1.863±0.121 |
| 5-8F/si-TRIM29 | 0.155±0.014 | 0.148±0.004*# | 0.287±0.037*# | 0.407±0.046*# | 0.777±0.076*# | 1.220±0.074*# | 1.380±0.062*# |
| F | 1.473 | 6.765 | 4.550 | 66.184 | 73.833 | 29.315 | 42.846 |
| P | 0.263 | 0.011 | 0.034 | 0.000 | 0.000 | 0.000 | 0.000 |
5-8F/si-TRIM29 vs 5-8F, P < 0.05;
5-8F/si-TRIM29 vs 5-8F/NC, P < 0.05.
Table 7. The absorbance values of 6-10B cells after transfecion (490 nm).
| cells | Day | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
| 6-10B | 0.144±0.009 | 0.155±0.041 | 0.252±0.021 | 0.432±0.027 | 0.660±0.034 | 1.099±0.085 | 1.437±0.092 |
| 6-10B/NC | 0.142±0.019 | 0.156±0.034 | 0.262±0.036 | 0.456±0.028 | 0.657±0.037 | 1.063±0.087 | 1.395±0.069 |
| 6-10B- TRIM29 | 0.148±0.017 | 0.170±0.022 | 0.285±0.038 | 0.570±0.064*# | 0.891±0.096*# | 1.497±0.062*# | 1.771±0.125*# |
| F | 0.211 | 0.342 | 1.548 | 17.266 | 19.051 | 50.934 | 17.903 |
| P | 0.812 | 0.716 | 0.247 | 0.000 | 0.000 | 0.000 | 0.000 |
6-10B-TRIM29 vs 6-10B, P < 0.05;
6-10B-TRIM29 vs 6-10B/NC, P < 0.05.
TRIM29, also known as Ataxia-telangiectasia group D complementing gene (ATDC), is involved in many human diseases, especially cancer [53]. TRIM29 expression in various tumor types has found that increased expression of TRIM29 is associated with more aggressive forms of disease including bladder [54], colorectal [55], gastric [56], lung [57], and pancreatic cancer [58]. TRIM29 expression in pancreatic cancer cells is 20 times higher than that in normal pancreas epithelial cells, overexpression of TRIM29 in pancreatic cancer lines promoted cell growth in vitro and metastatic activity in vivo stemming from stimulation of Wnt/β- Catenin/TCF signaling through TRIM29 binding to Dvl-2, a Wnt pathway activator downstream of the Frizzled receptor [19]. Another study found that TRIM29 functions as an oncogene in gastric cancer, the miR-185-TRIM29 axis regulates gastric cancer cell growth and apoptosis through Wnt/β-catenin pathway inactivation in vitro [59]. TRIM29 interacts with P53 and antagonizes p53-mediated functions in cancer cells, which increases cell proliferation via inhibition of p53 nuclear activities. TRIM29 represses the expression of p53-regulated genes, including p21 and NOXA. Mechanistically, TRIM29 binds p53, and this interaction is potentially fine-tuned by posttranslational acetylation of lysine 116 on TRIM29 [60]. TRIM29 can promote lung cancer proliferation through NF-kB induced up-regulation of cyclin D1 and c-Myc [61]. TRIM29 upregulates MMP-9 to promote lung cancer cell invasion by activating ERK and JNK pathways [62]. However, there is not report on the function of this protein in NPC. We found and confirmed that TRIM29 expression increased in NPC cells and tissues with high metastatic potentials, and high TRIM29 expression were correlated with lymph node and distant metastasis of PCR. To further investigate the effects of TRIM29 on NPC cell invasion and metastasis, we established NPC cell lines with stable TRIM29 knockdown or overexpression. We found that TRIM29 knockdown significantly attenuated while TRIM29 overexpression promoted NPC cell in vitro proliferation, migration and invasion and in vivo metastasis. Taken together, our results demonstrated that TRIM29 is a metastasis promoter protein of NPC. Furthermore, we will do further study of molecular mechanisms of NPC metastasis.
To get more insight on the biological significance of the differentially expressed proteins in NPC metastasis process, gene-ontology, KEGG pathways analysis and PPI were performed on 101 differential proteins. Cellular component, molecular functions, biological process which these proteins were enriched were performed by GO analysis. KEGG pathway analysis revealed that the differentially expressed proteins are involved in cancer-associated signaling pathways.
In summary, we identified a total of 101 DEPs in NPC cells with different metastatic potentials by iTRAQ-labeling combined with 2D-LC-MS/MS and found that the panel of the three proteins (RAN, SQSTM1 and TRIM29) could serve as novel potential biomarkers for predicting NPC metastasis, and high TRIM29 expression promoted NPC cell in vitro proliferation, migration and invasion and in vivo metastasis. These findings reported here could have potential clinical value in early warning NPC metastasis and also provide valuable information for further study of molecular mechanisms that govern NPC metastasis.
MATERIALS AND METHODS
Cell lines and tissue samples
High metastatic NPC 5-8F and non-metastatic NPC 6-10B cell lines were maintained in RPMI-1640 medium supplemented with 10% FBS in a humidified chamber with 5% CO2 at 37°C. The one hundred and eight formalin-fixed and paraffin-embedded archival NPC tissue specimens (64 NPCs with metastasis and 44 NPCs without metastasis) between Jan 2007 and Dec 2009 were obtained from the First Xiangya Hospital of Central South University and the First Hospital of Chenzhou City (Chenzhou, China) at the time of diagnosis before any therapy. On the basis of the 1978 WHO classification, all tumors were histopathologically diagnosed as poorly differentiated squamous cell carcinomas (WHO type III) [28]. The clinical stage of all the patients was classified according to the 1992 NPC staging system of China [29]. The clinicopathologic features of the patients used in the present study are shown in supplementary Table S-1.
Protein extraction, digestion and labeling with iTRAQ reagents
Cells were dissolved in lysis buffer (7 M urea, 2M thiourea, 65 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) at 4°C for 1 h, and then centrifuged at 12,000 rpm for 30 min at 4°C. The supernatant was collected, and the protein concentration was determined by 2D Quantification kit (Applied Biosystems). Trypsin digestion and iTRAQ labeling were performed according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). Briefly, 100μg protein of each sample was reduced and alkylated, then digested overnight at 37°C with trypsin (mass spectrometry grade; Promega, Madison, WI) and labeled with iTRAQ™ Reagents(Applied Biosystems) as follows: 6-10B, iTRAQ Reagent 121; 5-8F, iTRAQ Reagents 115 (the other channels for other samples). Two labeled digests were mixed and dried.
Off-line 2D-LC-MS/MS
The mixed peptides were fractionated by strong cation exchange chromatography on a 20AD HPLC system (Shimadzu) using a polySULFOETHYL column (2.1 100 mm, 5 m, 300Å; The Nest Group Inc.). Briefly, the mixed peptides were desalted with Sep-Pak Cartridge (Waters, Milford, MA), diluted with the loading buffer (10 mM KH2PO4 in 25% acetonitrile, pH 2.6), and loaded onto the column. Buffer A was identical in composition to the loading buffer, and buffer B was same as buffer A except that it contained 350 mM KCl. Separation was performed using a linear binary gradient of 0-88% buffer B in buffer A at a flow rate of 200μl/min for 60 min. The absorbance at 214 nm and 280 nm was monitored, and a total of 26 strong cation exchange fractions were collected along the gradient.
Each strong cation exchange fraction was dried down, dissolved in buffer C (5% acetonitrile, 0.1% formic acid), and analyzed on Qstar XL (Applied Biosystems). Briefly, peptides were separated on a reverse-phase (RB) column (ZORBAX 300SB-C18 column, 5 μm, 300Å, 0.1×15 mm; MicroMass) using a 20AD HPLC system (Shimadzu). The HPLC gradient was 5-35% buffer D (95% acetonitrile, 0.1% formic acid) in buffer C at a flow rate of 300 nL/min for 100 min. Survey scans were acquired from 400-1800 with up to four precursors selected for MS/MS from m/z 100-2000 using a dynamic exclusion of 30S. The iTRAQ labeled peptides fragmented under collision-induced dissociation conditions to give reporter ions at 115.1, 121.1 Th. The ratios of peak areas of the iTRAQ reporter ions reflect the relative abundances of the peptides and consequently, the proteins in the samples. Larger sequence-information-rich fragment ions were also produced under these MS/MS conditions and gave the identity of the protein from which the peptide originated. iTRAQ labeling followed by 2D-LC-MS/MS analysis was repeated twice(different protein lysates) to diminish the effect of experimental variation in the results of a proteomics analysis.
Data analysis
The software used for data acquisition was Analyst QS 1.1 (Applied Biosystems). The software used for protein identification and quantitation was ProteinPilot™ 4.2 (Applied Biosystems) with the integrated Paragon™ search algorithm and Pro Group™ algorithm (Revision Number: 3.0.0.0, 113442; Applied Biosystems). Identified proteins were grouped by the software to minimize redundancy. All the peptides used for the calculation of protein ratios were unique to the given protein or proteins within the group, and peptides that were common to other isoforms or proteins of the same family were ignored. The protein confidence threshold cutoff is 1.3 (unused ProtScore) with at least one peptide with 95% confidence. The average iTRAQ ratios from the twice experiments were calculated for each protein.
QRT-PCR
Total RNA were extracted from cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. 2 μg of total RNA was reversely transcribed for cDNA using the reverse transcription (RT) kit (Promega) and Oligo dT primer according to the manufacturer's instruction. The RT products were amplified by real-time PCR using QuantiFast SYBR green PCR kit (Qiagen) according to the manufacturer's instruction, and GAPDH was used as the internal control to normalize the expression levels of different genes. QRT-PCR was performed on the ABI Gene Amp PCR System 9700 (ABI). The primers used for the amplification of the indicated genes are listed in Supplementary Table S-2.
Western blotting
Cells were lysed in RIPA buffer. An equal amount of protein in each sample was mixed with Laemmli buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation, followed by blotting onto a polyvinylidene difluoride (PVDF) membrane (Millipore). Blots were incubated with primary anti-RAN antibody (1:1000; Proteintech), anti-TRIM29 antibody (1:200; Santa Cruz), anti-SQSTM1 antibody (1:100;Santa Cruz), or anti-actin antibody (1:2000; AURAGENE, China) overnight at 4°C, followed by incubation with goat anti-rabbit antibody (1:6000; Jackson Immuno Research) or goat anti-mouse antibody (1:3000; Jackson Immuno Research) for 1 h at room temperature. The signal was visualized with Luminata Crescendo Western HRP Substrate (Millipore, USA) and quantitated by densitometry using ImageQuant image analysis system (Storm Optical Scanner, Molecular Dynamics, Sunnyvale, CA).
Immunohistochemistry and evaluation of staining
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded tissue sections using a standard. Briefly, 4 μm of tissue sections were deparaffinized, rehydrated, and treated with an antigen retrieval solution (10 mmol/L sodium citrate buffer, pH 6.0). The sections were incubated with anti-RAN (1:100), anti-TRIM29 (1:50), or anti-SQSTM1 (1:50) antibody overnight at 4°C and then were incubated with 1:1000 dilution of biotinylated secondary antibody followed by avidin-biotin peroxidase complex (DAKO) according to the manufacturer's instructions. Finally, tissue sections were incubated with 3, 3′-Diaminobenzidine (Sigma) until a brown color was developed and counterstained with Harris modified hematoxylin. In negative controls, primary antibodies were omitted.
Immunostaining was blindly evaluated by two pathologists in an effort to provide a consensus on staining patterns by light microscopy. A quantitative score was performed by adding the score of staining extent and the score of staining intensity for each case to assess the expression levels of the proteins. Staining intensity was categorized: negative as 0, bordering as 1, weak as 2, moderate as 3, and strong as 4. The percentage of stained cells was categorized as 0–25% of stained cells = 1, 26–50% = 2, 51–75% = 3, and 76–100% =4. The staining score (ranging from 0-8) for each tissue was calculated by adding the area score and the intensity score. A combined staining score of ≤4 was considered to be low expression; and a score of >4 was considered to be high expression.
Establishment of NPC Cell Lines with overexpression and knockdown of TRIM29
To generate NPC cell lines with TRIM29 knockdown, GV102-TRIM29-shRNA vector and empty vector GV102 (Genechem, shanghai, China) were transfected into 5-8F cells using Lipofectamin 2000 (Invitrogen, USA) according to the manufacturer's instructions, respectively. To generate NPC cell lines with TRIM29 overexpression, GV143-TRIM29 expression vector and empty vector GV143 (Genechem) were transfected into the 6-10B cells using Lipofectamin 2000, respectively. Cells were selected using neomycin for 2 weeks, and NPC cell lines with stable overexpression and knockdown of TRIM29 and their respective control cell lines were obtained.
Cell proliferation assay
The cells in RPIM-1640 medium containing 10% fetal calf serum were plated at 2×103 cells per well in 96-well tissue culture plates, and grew for 6 days. Every 24 h, 20 μl of MTT (5 mg/ml; Sigma) was added to wells, and the medium was removed after 4 h of incubation. 250 μl of dimethylsulfoxide(DMSO) was added to each well for 10 min at room temperature. The absorbance of each well was read with a Bio-Tek Instruments EL310 Microplate Autoreader at 490 nm. MTT assay was performed three times in triplicate.
Wound healing assay
Cell migration was determined by scratch wound-healing assay. Briefly, cells were grown in RPMI 1640 medium containing 10% FBS overnight to confluence in a 6-well plate. Monolayers of cells were wounded by dragging a pipette tip. Cells were washed to remove cellular debris and allowed to migrate for 24-48h. Images were taken at 0, 24 and 48h after wounding under the inverted microscope.
Cell invasion assay
Invasion assays were performed in 24-well 8-mm pore size transwell chambers precoated with Matrigel (Corning) according to the manufacturer's instruction. The upper chamber was filled with 1×105 cells in RPMI 1640 medium containing 0.5% FBS. The lower chamber was filled with RPMI 1640 medium containing 10% FBS as a chemoattractant. After incubation at 37°C for 48h, cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Cells migrating through the Matrigel and the pores of the filter were counted from four random microscopic fields.
Experimental lung metastasis in nude mice
Nude male mice that were 4 weeks old were obtained from the Laboratory Animal Center of Central South University (Changsha, China) and were maintained under specific pathogen-free conditions. For experimental lung metastasis, mice (n = 6 each group) were injected intravenously with 1×107 cells/mouse via the tail vein. Six weeks after injection, mice were sacrificed by cervical dislocation, and lungs were removed, weighed, and embedded in paraffin for hematoxylin and eosin (H.E.) and immunohistochemical staining. Lung metastases were detected in paraffin-embedded tissue sections stained with H.E. staining.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software. Difference of RAN, SQSTM1, and TRIM29 protein expression between the two different kinds of tissues (metastatic NPC vs. non-metastatic NPC) was analyzed using Mann-Whitney U test. Moreover, the three proteins were individually, and as a panel, assessed for its ability to discriminate metastatic NPC from non-metastatic NPC lesions by evaluating its ROC curve based on the immunohistochemistry scores. Sensitivity, specificity, positive predictive value, and negative predictive value of the three proteins were calculated individually and as a panel. The relationship between RAN, SQSTM1, and TRIM29 expression and the clinicopathological characteristics in patients with NPC was analyzed using Mann-Whitney U test. Student's t-test was used for comparisons between two groups and one-way ANOVA was used for comparisons among more than two groups. A two-sided P < 0.05 was considered significant.
Ethics statement
This study was approved by the Institute Research Ethics Committee of Central South University, China. No informed consent (written or verbal) was obtained for use of retrospective tissue samples from the patients in the study, since most of the patients were deceased and informed consent was not deemed necessary and waived by the Ethics Committee. All animal experiments were undertaken in accordance with the Guide for the Care and Use of Laboratory Animals of Central South University, with the approval of the Scientific Investigation Board of Central South University.
Bioinformatics analysis
Differential expression proteins were annotated by GO using MAS 3.0 (http://bioinfo.capitalbio.com/mas3/). GO terms with computed p values less than 0.05 were considered as significantly enriched. KEGG pathway analysis was performed with the protein-protein interaction network of the differential proteins using Cytoscape (V2.8.2) with the ClueGO v1.4 plugin. A protein-protein interaction network(PPI) was constructed using Search Tool for the Retrieval of Interacting Genes/Proteins (String 9.05), in which the differential proteins served as a bait, and the proteins have a direct experimental interaction with the bait proteins in the databases. KEGG pathway was considered statistically significant when the corrected p value was less than 0.01.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
This study was supported by National Natural Science Foundation of China (81071796, 81172098), National Basic Research Program of China(2013CB910502), and Science and Technology Plan Projects of Changsha(k1406016-31).
Footnotes
CONFLICTS OF INTERESTS
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Adham M, Kurniawan AN, Muhtadi AI, Roezin A, Hermani B, Gondhowiardjo S, Tan IB, Middeldorp JM. Nasopharyngeal carcinoma in Indonesia: epidemiology, incidence, signs, and symptoms at presentation. Chin J Cancer. 2012;31:185–196. doi: 10.5732/cjc.011.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang ET, Adam HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 3.Siddique MA, Sabur MA, Kundu SC, Mostafa MG, Khan JA, Ahmed S, Karim MA, Hanif MA. Difficulty in diagnosis of nasopharyngeal carcinoma. Mymensingh Med J. 2012;21:158–161. [PubMed] [Google Scholar]
- 4.Liu J, Huang W, Ren CP, Wen QY, Liu WD, Yang XY, Wang L, Zhu B, Zeng L, Feng XL, Zhang C, Chen H, Jia W, Zhang LH, Xia XM, Chen YX. Flotillin-2 promotes metastasis of nasopharyngeal carcinoma by activating NF-B and PI3K/Akt3 signaling pathways. Sci Rep. 2015;5:11614. doi: 10.1038/srep11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JP, Zhan XQ, Li MY, Li GQ, Zhang PF, Xiao ZF, Shao MY, Peng F, Hu R, Chen ZC. Mitochondrial proteomics of nasopharyngeal carcinoma metastasis. BMC Med Genomics. 2012;5:62. doi: 10.1186/1755-8794-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melle C, Ernst G, Schimmel B, Bleul A, Koscielny S, Wiesner A, Bogumil R, Moller U, Osterloh D, Halbhuber KJ, von Eggeling F. A technical triade for proteomic identification and characterization of cancer biomarkers. Cancer Res. 2004;64:4099–4104. doi: 10.1158/0008-5472.CAN-03-3807. [DOI] [PubMed] [Google Scholar]
- 7.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 8.Ralhan R, Desouza V, Matta A, Chandra Tripathi S, Ghanny S, Datta Gupta S, Bahadur S, Siu KW. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteomics. 20087:1162–1173. doi: 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeSouza LV, Grigull J, Ghanny S, Dube V, Romaschin AD, Colgan TJ, Siu KW. Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:1170–1182. doi: 10.1074/mcp.M600378-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, Jaworska E, Lee CF, Blinco D, Okoniewsk MJ, Miller CJ, Bitton DA, Spooncer E, Whetton AD. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics. 2008;7:853–863. doi: 10.1074/mcp.M700251-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Feng X, Zhang J, Chen WN, Ching CB. Proteome profiling of Epstein-Barr virus infected nasopharyngeal carcinoma cell line: identification of potential biomarkers by comparative iTRAQ-coupled 2D LC/MS-MS analysis. J Proteomics. 2011;74:567–576. doi: 10.1016/j.jprot.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Z, Li G, Chen Y, Li M, Peng F, Li C, Li F, Yu, Ouyang Y, Xiao Z, Chen Z. Quantitative proteomic analysis of formalin-fixed and paraffin- embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J Histochem Cytochem. 2010;58:517–527. doi: 10.1369/jhc.2010.955526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LC, Chung IC, Hsueh C, Tsang NM, Chi LM, Liang Y, Chen CC, Wang LJ, Chang YS. The antiapoptotic protein, FLIP, is regulated by heterogeneous nuclear ribonucleoprotein K and correlates with poor overall survival of nasopharyngeal carcinoma patients. Cell Death Differ. 2010;17:1463–1473. doi: 10.1038/cdd.2010.24. [DOI] [PubMed] [Google Scholar]
- 14.Mo Y, Hou H, Li D, Liang Y, Chen D, Zhou Y. Mitochondrial protein targets of radiosensitisation by 1,8-dihydroxy-3-acetyl-6-methyl-9,10 anthraquinone on nasopharyngeal carcinoma cells. Eur J Pharmacol. 2014;738:133–141. doi: 10.1016/j.ejphar.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 15.Cai XZ, Zeng WQ, Xiang Y, Liu Y, Zhang HM, Li H, She S, Yang M, Xia K, Peng SF. iTRAQ-based Quantitative Proteomic Analysis of Nasopharyngeal Carcinoma. J Cell Biochem. 2015;116:1431–1441. doi: 10.1002/jcb.25105. [DOI] [PubMed] [Google Scholar]
- 16.Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R, Aiken C, Charlton LM, Gronenborn AM, Zhang P. Rhesus TRIM5alpha disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7:e1002009. doi: 10.1371/journal.ppat.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira NM, Trikha R, McKnight A. A novel envelope mediated post entry restriction of murine leukaemia virus in human cells is Ref1/TRIM5alpha independent. Retrovirology. 2010;7:81. doi: 10.1186/1742-4690-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L, Fearon ER, Ljungman M, Simeone DM. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and betacatenin stabilization. Cancer Cell. 2009;15:207–219. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawthorn L, Stein L, Panzarella J, Loewen GM, Baumann H. Characterization of cell type specific profiles in tissues and isolated cells from squamous cell carcinomas of the lung. Lung Cancer. 2006;53:129–142. doi: 10.1016/j.lungcan.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Kosaka Y, Jnoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- 22.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Møller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 23.Glebov OK, Rodriguez LM, Soballe P, DeNobile J, Cliatt J, Nakahara K, Kirsch IR. Gene expression patterns distinguish colonoscopically isolated human aberrant crypt foci from normal colonic mucosa. Cancer Epidemiol Biomarkers Prev. 2006;15:2253–2262. doi: 10.1158/1055-9965.EPI-05-0694. [DOI] [PubMed] [Google Scholar]
- 24.Ohmachi T, Tanaka F, Mimori K, Inoue H, Yanaga K, Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin Cancer Res. 2006;12:3057–3063. doi: 10.1158/1078-0432.CCR-05-1961. [DOI] [PubMed] [Google Scholar]
- 25.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy J., Jr Pecorelli S. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 26.Mutter GL, Baak JP, Fitzgerald JT, Gray R, Neuberg D, Kust GA, Gentleman R, Gullans SR, Wei LJ, Wilcox M. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol Oncol. 2001;83:177–185. doi: 10.1006/gyno.2001.6352. [DOI] [PubMed] [Google Scholar]
- 27.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, Anaissie E, Morris C, Muwalla F, van Rhee F, Fassas A, Crowley J, Tricot G, Barlogie B, Shaughnessy J., Jr Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 28.Shanmugaratnam K, Sobin LH. The World Health Organization histological classification of tumours of the upper respiratory tract and ear. A commentary on the second edition. Cancer. 1993;71:2689–2697. doi: 10.1002/1097-0142(19930415)71:8<2689::aid-cncr2820710843>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 29.Qian CN, Guo X, Cao B, Kort EJ, Lee CC, Chen J, Wang LM, Mai WY, Min HQ, Hong MH, Vande Woude GF, Resau JH, Teh BT. Met protein expression level correlates with survival in patients with late-stage nasopharyngeal carcinoma. Cancer Res. 2002;62:589–596. [PubMed] [Google Scholar]
- 30.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 31.Yuen HF, Chan KK, Grills C, Murray JT, Plat-Higgins A, Eldin OS, O'Byrne K, Janne P, Fennell DA, Johnston PG, Rudland PS, El-Tanani M. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res. 2012;18:380–391. doi: 10.1158/1078-0432.CCR-11-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Zhao X, Li K, Luo G, Nie Y, Shi Y, Zhou Y, Ren G, Feng B, Liu Z, Pan Y, Li T, Guo X, Wu K, Miranda-Vizuete A, Wang X, Fan D. Thioredoxin-like protein 2 is overexpressed in colon cancer and promotes cancer cell metastasis by interaction with ran. Antioxid Redox Signal. 2013;19:899–911. doi: 10.1089/ars.2012.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe H, Kamai T, Shirataki H, Oyama T, Arai K, Yoshida K. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer. 2008;122:2391–2397. doi: 10.1002/ijc.23400. [DOI] [PubMed] [Google Scholar]
- 34.Azuma K, Sasada T, Takedatsu H, Shomura H, Koga M, Maeda Y, et al. Ran, a small GTPase gene, encodes cytotoxic T lymphocyte (CTL) epitopes capable of inducing HLA-A33-restricted and tumor-reactive CTLs in cancer patients. Clin Cancer Res. 2004;10:6695–6702. doi: 10.1158/1078-0432.CCR-04-0818. [DOI] [PubMed] [Google Scholar]
- 35.Kurisetty V, Johnston P, Johnston N, Erwin P, Crowe P, Fernig D, et al. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:7139–7149. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 36.Ly TK, Wang J, Pereira R, Rojas KS, Peng X, Feng Q, et al. Activation of the Ran GTPase is subject to growth factor regulation and can give rise to cellular transformation. J Biol Chem. 2010;285:5815–5826. doi: 10.1074/jbc.M109.071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan H, Lu Y, Qin H, Zhou Y, Gu Y, Zhou J, Wang X, Fan D. High Ran level is correlated with poor prognosis in patients with colorectal cancer. Int J Clin Oncol. 2013;18:856–863. doi: 10.1007/s10147-012-0465-x. [DOI] [PubMed] [Google Scholar]
- 38.Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J Biol Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- 39.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem. 2005;280:35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 42.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphory- lation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 43.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, Liu J, Lemischka IR, Hung MC, Chiao PJ. KrasG12D-induced IKK2/beta/NF- kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, Dinsdale D, Condorelli F, Brandner S, Campanella M, Grose R, Jones C, Salomoni P. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 47.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trocoli A, Bensadoun P, Richard E, Labrunie G, Merhi F, Schlafli AM, Brigger D, Souquere S, Pierron G, Pasquet J-M, Soubeyran P, Reiffers J, Segal-Bendirdjian E, Tschan MP, Djavaheri-Mergny M. p62/SQSTM1 upregulation constitutes a survival mechanism that occurs during granulocytic differentiation of acute myeloid leukemia cells. Cell Death and Differentiation. 2014;21:1852–1861. doi: 10.1038/cdd.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino o, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee M S, Tanaka K, Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, DiPaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ, Brakenhoff RH. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin Cancer Res. 2013;19:1994–2003. doi: 10.1158/1078-0432.CCR-12-2539. [DOI] [PubMed] [Google Scholar]
- 54.Fristrup N, Birkenkamp-Demtroder K, Reinert T, Sanchez-Carbayo M, Segersten U, Malmstrom PU, Palou J, Alvarez-Múgica M, Pan CC, Ulhøi BP, Borre M, Ørntoft TF, Dyrskjøt L. Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer. Am J Pathol. 2013;182:339–349. doi: 10.1016/j.ajpath.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Glebov OK, Rodriguez LM, Soballe P, DeNobile J, Cliatt J, Nakahara K, Kirsch IR. Gene expression patterns distinguish colonoscopically isolated human aberrant crypt foci from normal colonic mucosa. Cancer Epidemiol Biomarkers Prev. 2006;15:2253–2262. doi: 10.1158/1055-9965.EPI-05-0694. [DOI] [PubMed] [Google Scholar]
- 56.Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- 57.Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, Back T, Song NY, Datla M, Sun Z, Goldszmid R, Lin F, Cohoon T, Pike K, Wu X, Schrump DS, Wong KK, Young HA, Trin chieri G, Wiltrout RH, Hu Y. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell. 2013;23:527–540. doi: 10.1016/j.ccr.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 59.Qiu Feng, Jian-Ping Xiong, Jun Deng, Xiao-Jun Xiang. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. 2015;8:5053–5061. [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan Z, Villagra A, Peng L, Coppola D, Glozak M, Sotomayor EM, Chen J, Lane WS, Seto E. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. 2010;30:3004–3015. doi: 10.1128/MCB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang ZP, Dong QZ, Cui QZ, Papavassiliou P, Wang ED, Wang EH. Ataxia- Telangiectasia Group D Complementing Gene (ATDC) Promotes Lung Cancer Cell Proliferation by Activating NF-kB Pathway. PLOS ONE. 2013;8:e63676. doi: 10.1371/journal.pone.0063676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang ZP, Cui QZ, Dong QZ, Xu K, Wang EH. Ataxia-telangiectasia group D complementing gene (ATDC) upregulates matrix metalloproteinase 9 (MMP-9) to promote lung cancer cell invasion by activating ERK and JNK pathways. Tumor Biol. 2013;34:2835–2842. doi: 10.1007/s13277-013-0843-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


