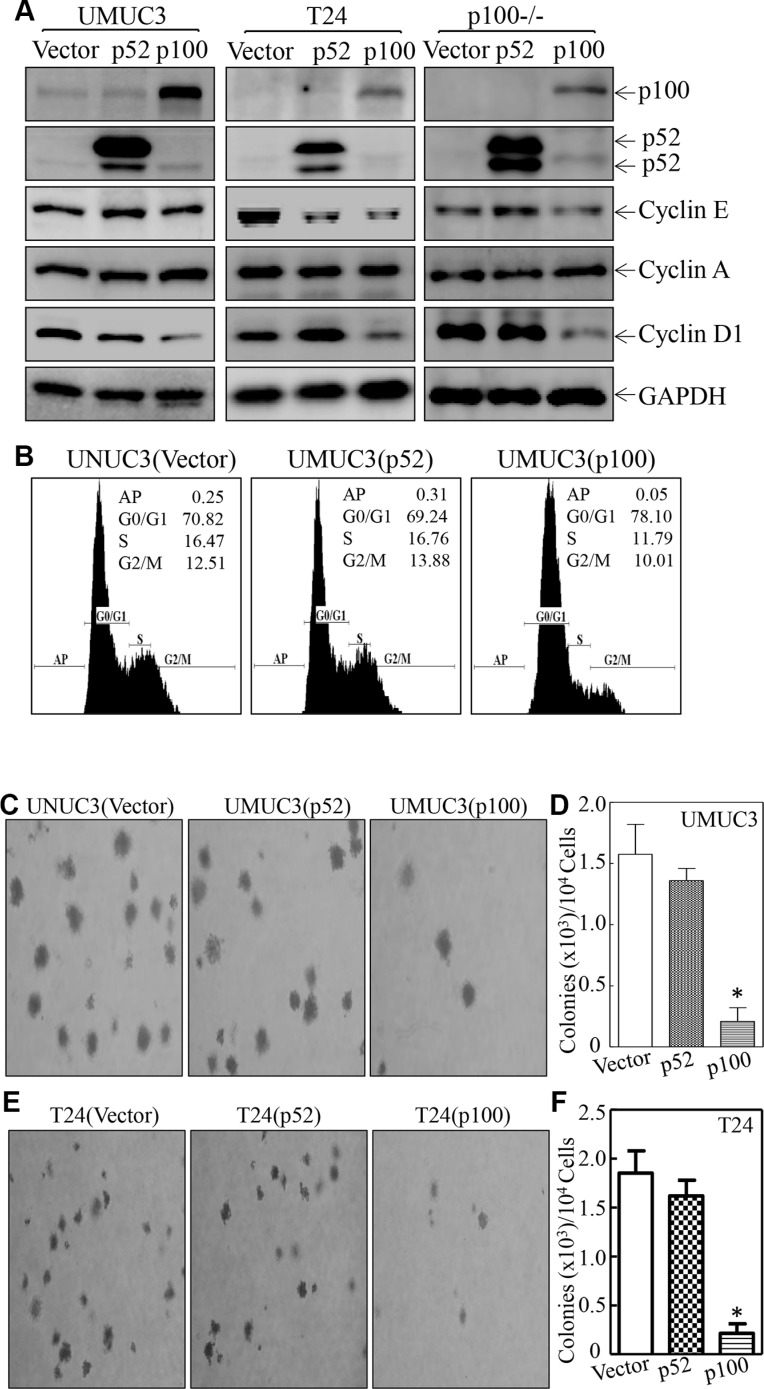
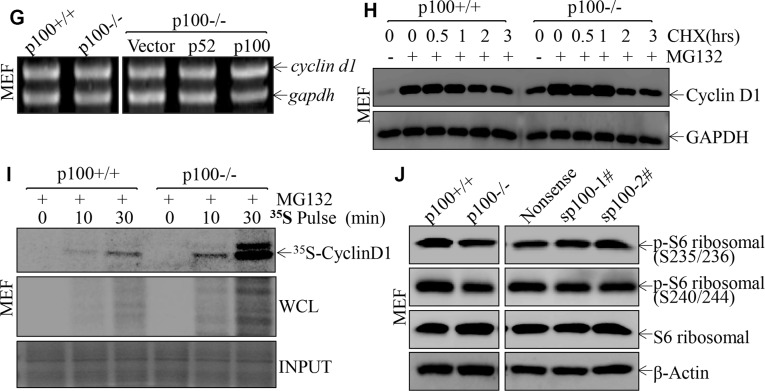
Figure 3. NFκB2 p100, but not p52, mediated the inhibition of Cyclin D1 expression, cell cycle progression and anchorage-independent growth in UMUC3 and T24 cells.
(A) The cell extracts were subjected to Western Blot as indicated. GAPDH was used as a loading control. (B) Flow-cytometry analysis of cell cycle alteration was performance as indicated. (C–F) Anchorage-independent growth was determined in soft agar assay as indicated. The number of colonies was scored and presented as colonies per ten thousand cells. The symbol (*) indicates a significant decreases in comparison to the control transfectant (p < 0.05). (G) Total RNA was isolated from the indicated cells and then subjected to RT-PCR analysis of cyclin d1 mRNA expression. The gapdh was used as a loading control. (H) After pretreatment with MG132 (10 μM) for 4 h, p100+/+ and p100−/− cells were subjected to determination of Cyclin D1 protein degradation in the presence of cycloheximide (CHX) (50 μM). GAPDH was used as a loading control. (I) After pre-treatment with MG132 (10 μM) for 30 mins, newly synthesized Cyclin D1 protein in p100+/+ and p100−/− cells was monitored by pulse assay using 35S-labeled methionine/cysteine. WCL stands for whole cell lysate. Coomassie blue staining was used for protein loading control. (J) The cell extracts were subjected to Western Blot as indicated. β-Actin was used as a loading control.