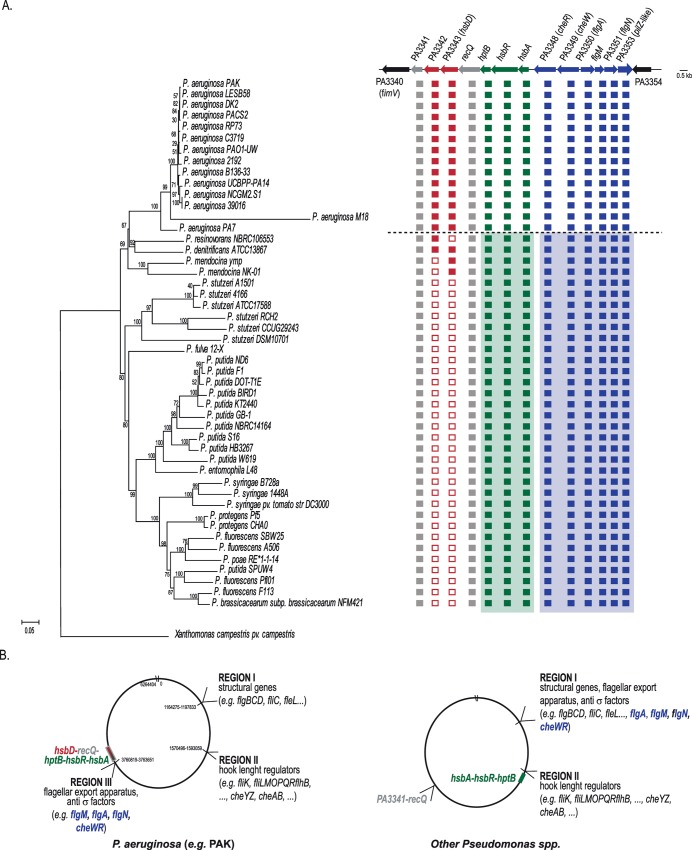
Fig 1. Occurrence of PA3343 (hsbD) in Pseudomonas strains and its relation with the flagellar genes reorganization in P. aeruginosa.
(A) Evolutionary relationships of Pseudomonas taxa carrying hptB orthologs and taxonomic distribution of PA3341-PA3353 genes. The phylogenetic tree of Pseudomonas strains was constructed using MEGA 6 [58]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Filled squares indicate the presence of an ortholog gene, while empty squares indicate the absence. Location of the PA3341-PA3353 genes is shown as in P. aeruginosa. When a green or blue background is used, it indicates that each gene set is present, but at different location on the chromosome, as explained in panel B. Non-aeruginosa strains are separated by a dashed line. (B) Location of flagella and hptB related genes in Pseudomonas. In P. aeruginosa (left) flagella genes are located in three regions of the chromosome [68] while in other Pseudomonas species (right) two regions are present and in general physically separated with some exceptions (e.g. Pseudomonas entomophila L48) [26].