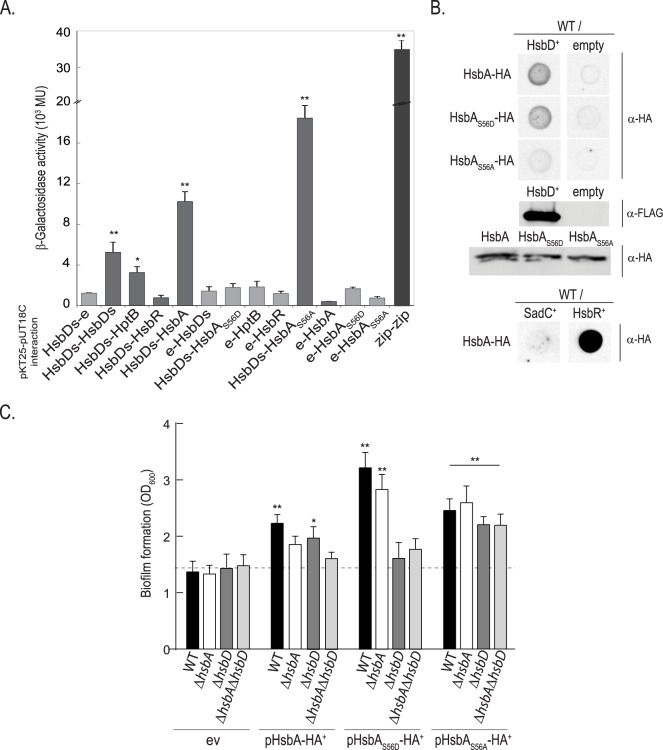
Fig 3. Interaction of HsbD with HsbA is dependent on HsbA phosphorylation.
(A) Reconstitution of adenylate cyclase in the E. coli strain DHM1 using a bacterial two-hybrid approach (see S1 Table) was detected by blue staining due to X-gal hydrolysis when colonies where grown on LB–X-gal agar plates containing 0.5 mM IPTG, 100 μg/ml ampicillin, and 50 μg/ml chloramphenicol agar plates. The interactions were also quantified by β-galactosidase assays using liquid cultures of the same strains. Each value is the average of three different cultures ± standard deviation (*, p < 0.05; **, p < 0.01). (B) Dot blot analysis of HsbD-HsbA interaction in P. aeruginosa PAK. PAK wild type (WT) cell lysates overexpressing HsbD (FLAG-tagged), SadC, HsbR (prey proteins) are spotted on a membrane and incubated with purified HA-tagged HsbA, HsbAS56D and HsbDS56A proteins (bait proteins). Detection of HsbA bait proteins bound to HsbD prey on the blot was performed using α-HA antibody. Empty vector was used as negative control. Production of HsbD-FLAG/HsbA-HA variants is shown by Western blot. (C) Biofilm formation of P. aeruginosa wild-type PAK, ΔhsbA, ΔhsbD and ΔhsbAΔhsbD strains measured by crystal violet staining. Bacterial strains were expressing in trans either HsbA, HsbAS56D or HsbAS56A and grown in LB medium in 24-microtiter plates for 14 hours in presence of 50 μg/ml gentamycin and 0.5 mM IPTG. ev: empty vector (pME6032, see S1 Table). Each value is the average of three different cultures ± standard deviation. Asterisks indicate statistically significant difference of biofilm formation compared to the ev (*, p< 0.05; **, p< 0.01).