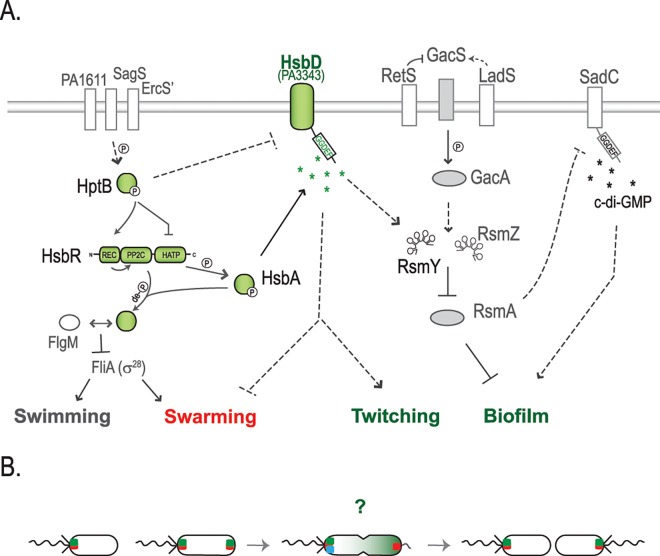
Fig 11. Working model for the HptB signaling pathway in P. aeruginosa.
(A) HptB is activated (i.e. phosphorylated) via either one of three orphan sensor kinase hybrids: PA1611, ErcS’ (PA1976) or SagS (PA2824) [26–28]. When phosphorylated, HptB transfers a phosphoryl group to the HsbR receiver domain (HsbR-P), thus repressing HsbR kinase activity while activating its phosphatase activity. HsbR-P dephosphorylates the anti-anti-sigma factor HsbA. Dephosphorylated HsbA leads to dissociation of the HsbA-HsbR complex and a subsequent stable sequestration of the anti-sigma factor FlgM by HsbA [20]. The interaction HsbA-FlgM induces flagellar genes expression, by allowing the release of the FliA sigma factor (σ28) [51]. When HptB is inactive (dephosphorylated or in an hptB mutant), swimming and swarming are no longer supported. Instead, the HsbR kinase phosphorylates HsbA (HsbA-P) and HsbA-P interaction with HsbD leads to an increase of c-di-GMP and RsmY levels. The activation of HsbD strengthens swarming repression and results in hyper-biofilm and hyper-twitching phenotypes. This cascade of events is in agreement with the waves of regulatory actions that cause bacteria progression from an early surface attachment and colonization (swimming and swarming) to the development of mature sessile biofilms [15, 26, 56]. The symbol ➔ indicates positive regulation, while -¦ repression and de–P (de)phosphorylation. Dashed lines suggest a probable indirect regulation. Components of the HptB signaling pathway are colored in green. (B) Localization dynamics of HsbD (green square) during cell division. FhlF (red square) dynamics is reported as commented previously by Burrows LL [49]. Green gradient in the cell undergoing division illustrates bimodal distribution of c-di-GMP levels as reported by Christen and colleagues [14].? Question mark represents possible scenarios on the asymmetric partitioning of HsbD during cell division in respect to flagellum biogenesis and DipA (blue square) localization. For details see text.