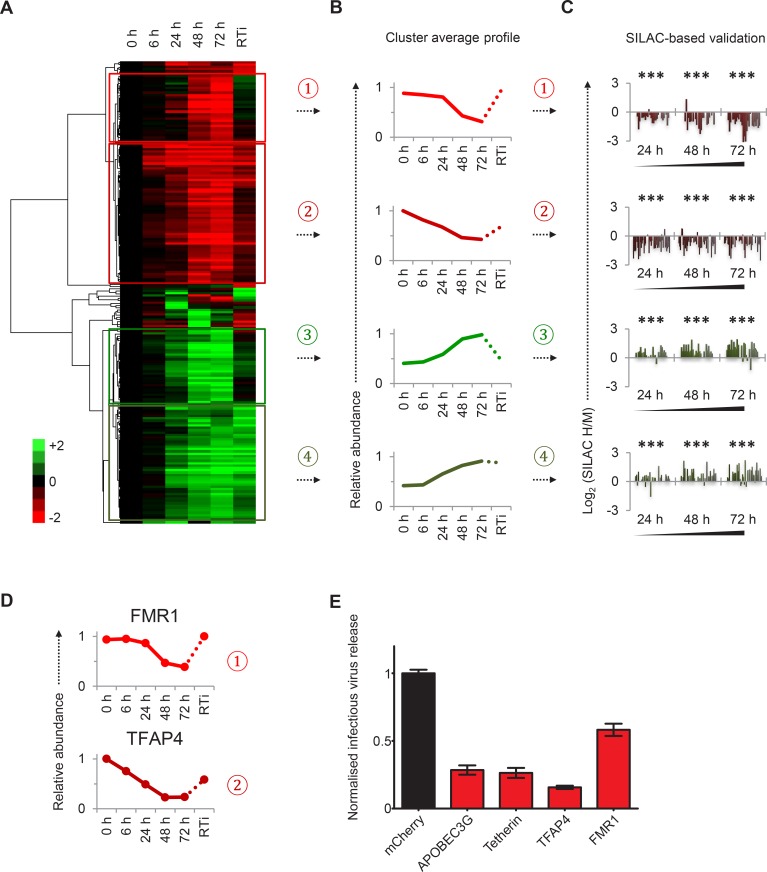
Figure 2. Identification and SILAC-based proteomic validation of novel HIV targets.
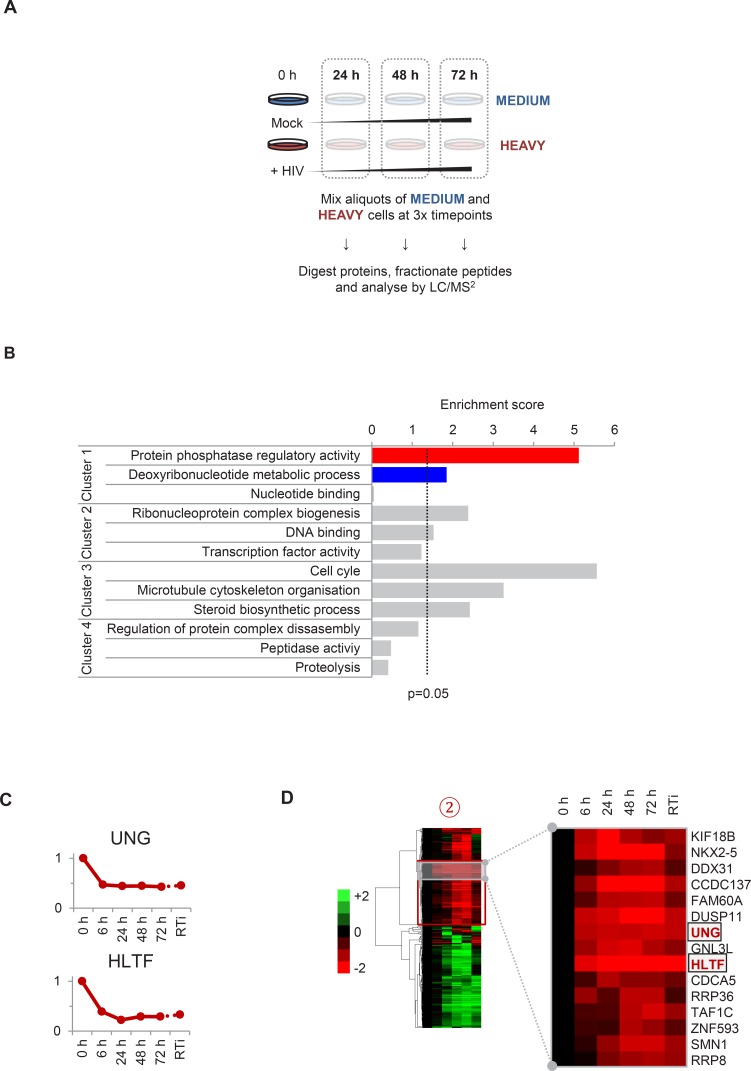
(A) Hierarchical cluster analysis of temporal profiles of proteins regulated by HIV. The heatmap shows log2 (fold change in protein abundance compared with uninfected cells) and clusters #1–4 are indicated. (B) Average temporal profiles of proteins in clusters #1–4. Relative abundance is expressed as a fraction of maximum TMT reporter ion intensity. (C) SILAC-based validation of novel HIV targets. CEM-T4 T cells pre-labelled with heavy amino acids were infected with NL4-3-dE-EGFP HIV at an MOI of 10, and control cells pre-labelled with medium amino acids were mock-infected without virus. Aliquots of HIV-infected (heavy; H) and mock (medium; M) cells were harvested sequentially at the indicated timepoints and subjected to SILAC-based whole cell proteomic analysis (Figure 2—figure supplement 1A). Log2 (H/M protein abundance) at 24, 48 and 72 hr is shown for proteins from clusters #1–4. ***p value<0.001. (D) Temporal profiles of novel HIV-1 targets FMR1 and TFAP4. Relative abundance is expressed as a fraction of maximum TMT reporter ion intensity. (E) Antagonism of HIV production by FMR1 and TFAP4. 293T cells were co-transfected with pNL4-3-dE-EGFP/pMD.G and either mCherry or the indicated cellular protein. 48 hr culture supernatants were assayed for infectious virus by infection of HeLa cells. Well-characterised restriction factors APOBEC3G and tetherin were included as controls, and infectious virus release normalised compared with mCherry. Mean values and standard errors are shown from at least 4 replicates. All four proteins significantly reduced viral release compared with mCherry in an ANOVA analysis with Bonferroni post-test, p values<0.001.
DOI: http://dx.doi.org/10.7554/eLife.18296.007