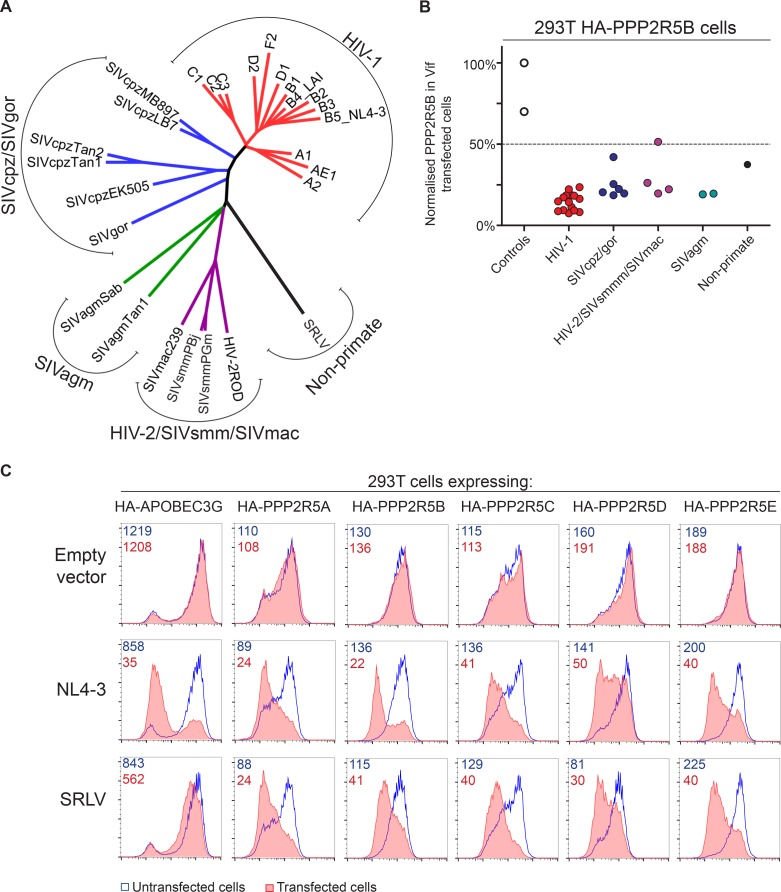
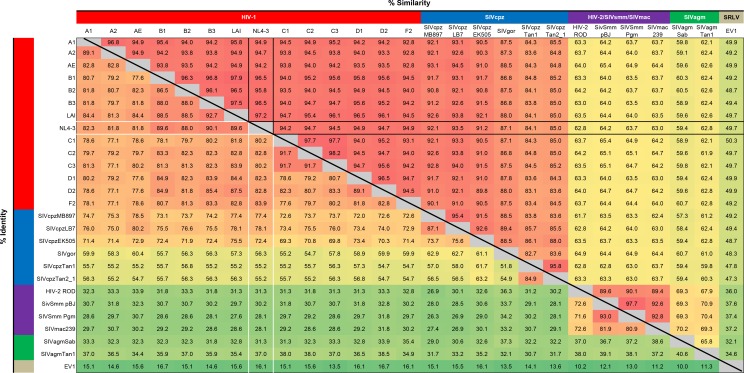
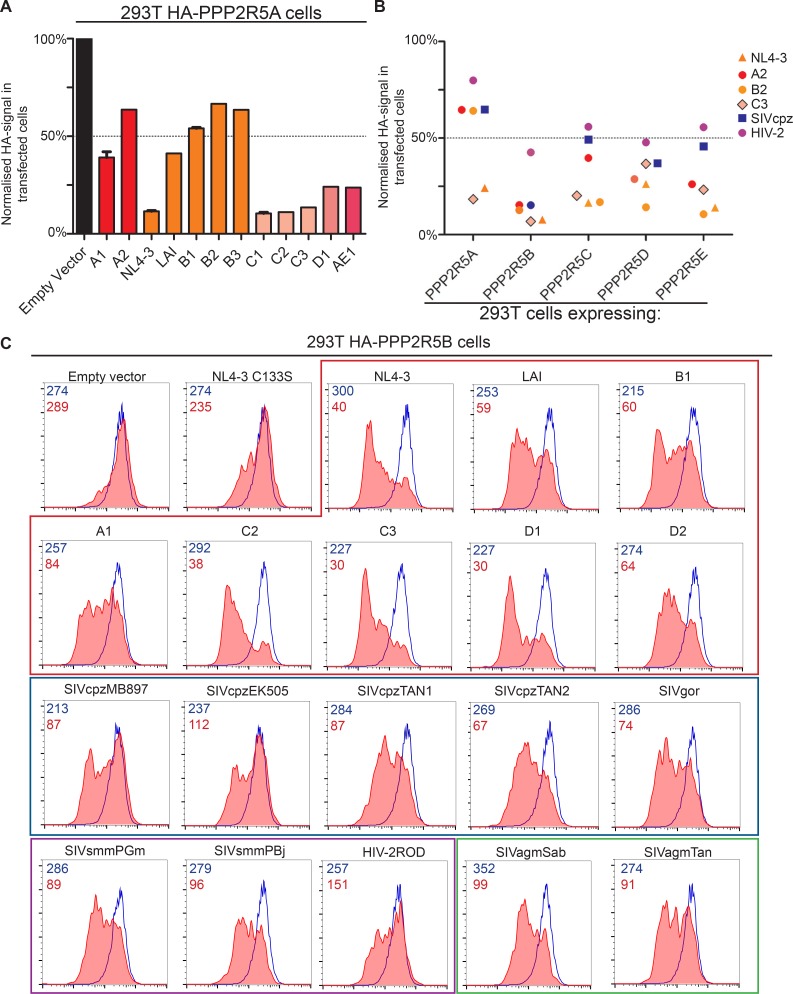
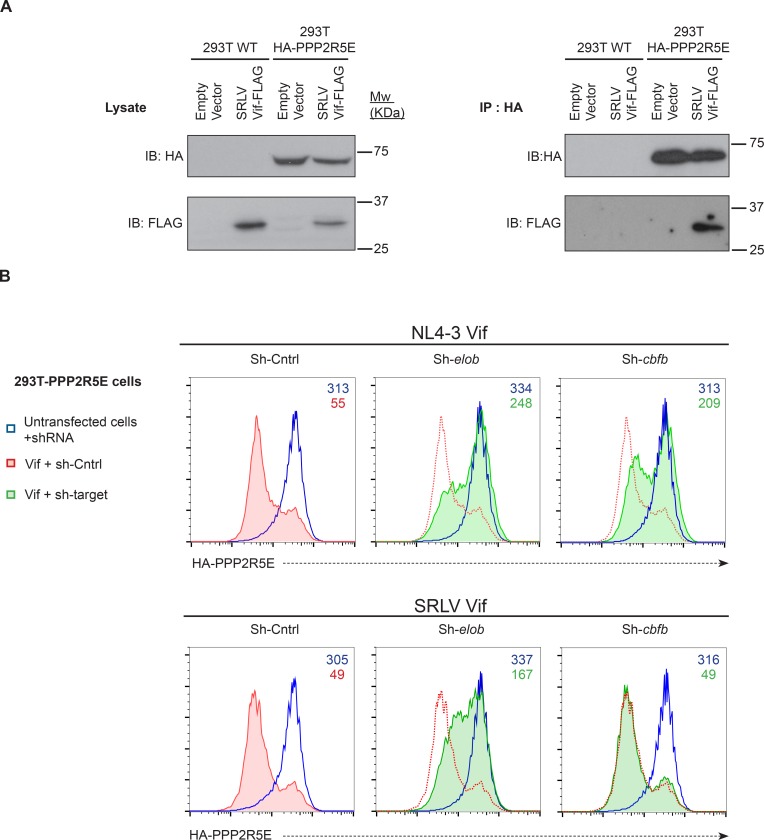
Figure 7. Phylogenetic conservation of PPP2R5A-E subunit degradation.
(A) Phylogenetic tree based on the amino acid alignment of Vif variants used in this study. (B) Conservation of PPP2R5B subunit degradation by phylogenetically diverse lentiviral Vif variants. 293T cells stably expressing HA-PPP2R5B were transfected with a panel of lentiviral Vif variants and analysed by intracellular flow cytometry for HA 36 hr post-transfection. The median fluorescence intensity of the transfected population is shown as a proportion of median fluorescence intensity of the untransfected population for each condition, normalized to the empty vector control. Datapoints represent mean values for different Vif variants obtained from up to four independent experiments. (C) Depletion of PPP2R5A-E subunits by small ruminant lentivirus Vif. 293T cells stably expressing HA-PPP2R5A-E or HA-APOBEC3G were transfected with NL4-3 (HIV-1) or SRLV Vif variants. Histograms show GFP positive (transfected, red shading) and negative (untransfected, blue line) cells. MFI values are shown for GFP positive (red) and negative (blue) cells.