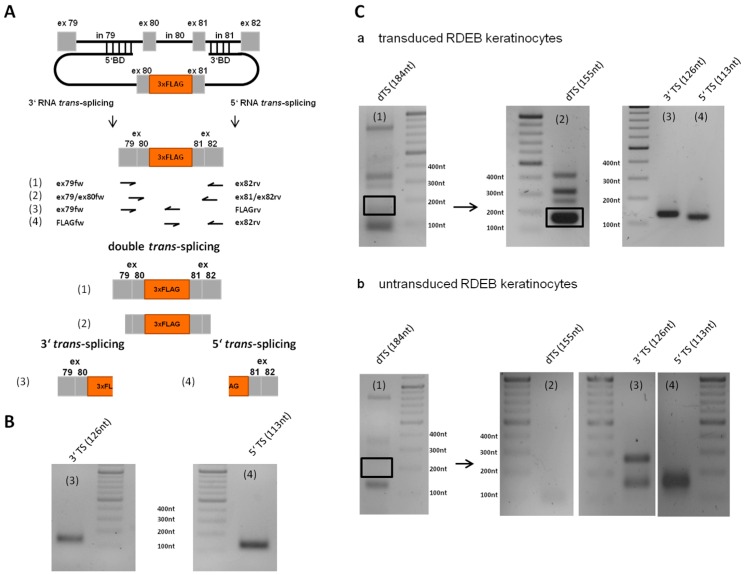
Figure 7.
Detection of endogenous double trans-splicing in an RDEB patient cell line. (A) Schematic depiction of the endogenous double trans-splicing analysis system. dRTM15 contains the BDs for 3′ and 5′ trans-splicing induction, preselected using a COL7A1-MG-based dRTM screening system, and the sequence-optimized wild-type coding sequence of exon 80 and 81 interrupted by a 3xFLAG tag sequence. In order to detect accurate 3′, 5′ and double trans-splicing on endogenous RNA level different primer sets were used for the respective PCR amplification (1–4); (B) Detection of 3′ trans-splicing (126 nt) and 5′ trans-splicing (113 nt) separately. trans-splicing products (3, 4) were confirmed by Sanger sequence analysis; (C) (a) The expected area on the agarose gel, at which the 184 nt long double RNA trans-splicing product (1), was excised from the gel, purified and included as a template for subsequently performed nested PCR, revealing accurate 3′ (3), 5′ (4) and double trans-splicing (2) into endogenous COL7A1 transcripts (3′ trans-splicing: 126 nt, 5′ trans-splicing: 113 nt, double trans-splicing: 155 nt); and (b) as negative control cDNA of untransduced patient keratinocytes was used as template and the PCRs were performed as preceded in (a). Unspecific PCR products were excised from the gel, purified and confirmed by Sanger sequence analysis as endogenous COL7A1 sequences without RTM sequence.