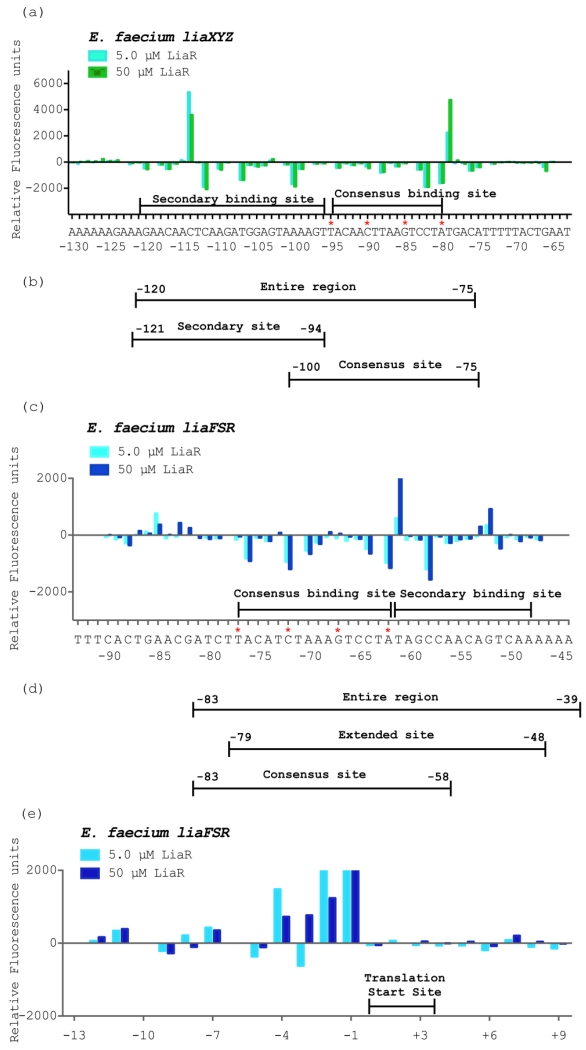
Figure 5. Efm LiaR recognizes complex upstream regulatory regions that extend beyond the predicted consensus sequence.
DNase I Footprinting followed by Automated Capillary Electrophoresis (DFACE) was used to establish the Efm LiaR binding footprint within the upstream regions of the liaXYZ (−292 to +65) and liaFSR (−281 to +98) operons of E. faecium R494. Protection studies were performed at 0.5, 5 and 50 μM LiaR activated with the BeF3−. DNaseI digestion patterns are shown as histograms (a, c, e) where negative changes in relative fluorescence units indicate regions of protection and positive changes indicate hypersensitivity. All DNaseI sensitivity data are relative to a no protein negative control.
(a) Efm LiaR binding to the promoter region of the liaXYZ operon. Nucleotide positions refer to the region of LiaR binding (−120 to −75) on the DNA relative to the translation start site of LiaX (cyan, 5 μM LiaR; green, 50 μM LiaR).
(b/d) Oligonucleotides used for DNA binding studies (Table 2).
(c) Efm LiaR binding to the promoter region of the liaFRS operon. Nucleotide positions refer to the region of LiaR binding (−77 to −48) on the DNA relative to the translation start site of LiaF (cyan, 5 μM LiaR; blue, 50 μM LiaR).
(e) Superimposed electropherograms showing a strong and consistent hypersensitive site at positions −4 to −1 (AGCG) relative to the translation start site of LiaF in the presence of Efm LiaR (cyan, 5 μM LiaR; blue, 50 μM LiaR).