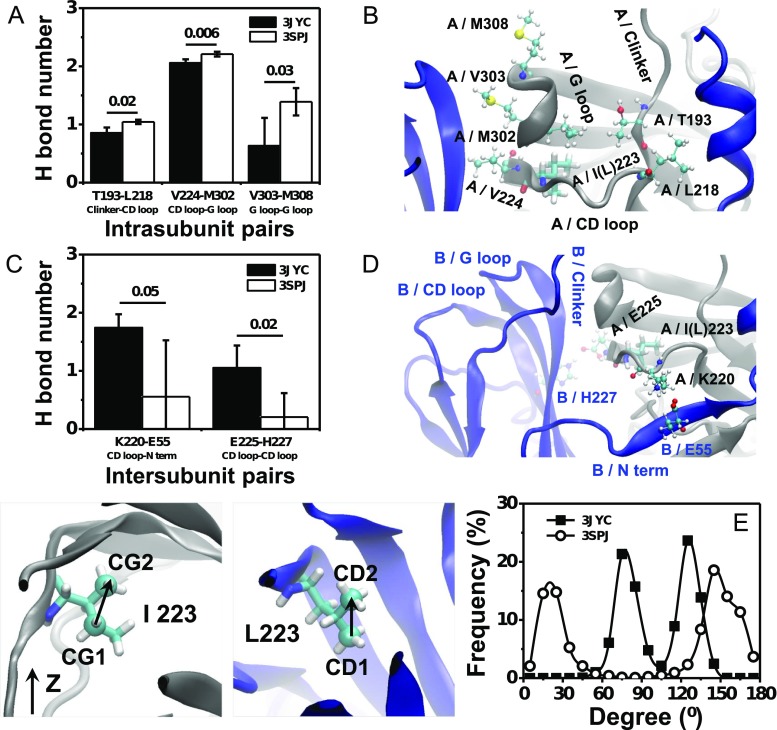
Fig. 4.
Difference in the interaction networks of the PIP2-absent WT-3JYC and I223L-3SPJ mutant of chicken Kir2.2. Key amino acid pairs of the intrasubunit interaction with significantly different H bond interactions (a) and the corresponding location (b) are shown. Those of the intersubunit interaction are presented in c, d. The distributions of the sidechain orientation of I223 and L223 are shown in e with a bin size of 15°. The H bond interactions are presented as the mean ± SD of the tetramer after the last 10-ns trajectory average of each subunit. The key amino acids are presented as CPK with labeling of the subunit and the corresponding secondary structures. I(L)223 is shown in Licorice for clarity