Abstract
Background
Patients with rectal cancer undergo preoperative neoadjuvant chemoradiation with approximately 70% exhibiting pathologic down-staging in response to treatment. There is currently no accurate test to predict patients who are likely to be complete responders to therapy. 5-Fluorouracil (5-FU) is regularly used in the neoadjuvant treatment of rectal cancer. Genetic polymorphisms affect the activity of thymidylate synthase (TS), an enzyme involved in 5-FU metabolism. This may account for observed differences in response to neoadjuvant treatment between patients. Detection of such polymorphisms might identify patients likely to have a complete response to neoadjuvant therapy and perhaps allow them to avoid surgery.
Methods
DNA was isolated from whole blood taken from patients with newly diagnosed rectal cancer who received neoadjuvant therapy (n=50). Response to therapy was calculated with a tumor regression score, based upon histology, from the time of surgery. PCR was performed targeting the promoter region of TS. PCR products were separated using electrophoresis to determine whether patients were homozygous for a double-tandem repeat (2R), a triple-tandem repeat (3R), or heterozygous (2R/3R). A single nucleotide polymorphism (SNP), 3G or 3C, may also be present in the second repeat unit of the 3R allele. Restriction fragment length polymorphism assays were performed in patients with at least one 3R allele using HaeIII.
Results
Patients with at least one TS 3G allele were more likely to have complete or partial pathological response to 5-FU neoadjuvant therapy (OR 10.4 95% CI 1.3–81.6) (p=0.01) than those without at least one 3G allele.
Conclusions
Identification of patients with specific genetic polymorphisms in enzymes involved in 5-FU metabolism appears to predict the likelihood of complete or partial pathologic response of rectal cancer to preoperative neoadjuvant therapy.
Introduction
In the United States, colorectal cancer is the third most common cancer in men and women, as well as the third leading cause of cancer death (1). Studies have demonstrated the usefulness of neoadjuvant chemotherapy and radiation for locally advanced local rectal cancer (2). The benefits of preoperative neoadjuvant chemotherapy in rectal cancer include tumor downstaging and complete pathologic response or total destruction of the cancer by such therapy. Patients with complete pathologic response after neoadjuvant chemotherapy have better outcomes than those without complete response (3).
The key to successful neoadjuvant treatment is selecting the best-suited patients. Pharmacogenomics is a field that combines pharmacology and genetics to assure that patients are receiving personalized therapy or therapy best-suited to their particular disease (4). Physicians can use patient genotypes to guide pharmacological treatment, minimize adverse drug effects and toxicities, and maximize treatment outcomes (5). Single Nucleotide Polymorphisms (SNPs) are one of the genetic tools used in pharmacogenomics and are single nucleotide variations within a DNA sequence (5). We believe that it can assist in selection of which rectal cancer patients would benefit most from preoperative chemoradiation.
5-Fluorouracil (5-FU) is a pyrimidine analog that is a mainstay drug used for preoperative neoadjuvant chemoradiation (nCRT) in patients with locally advanced rectal cancer (6). Approximately 70% of patients exhibit pathologic downstaging following treatment, 15–20% of patients exhibit complete pathologic response, while 10–15% of patients have no pathologic response to 5-FU treatment (7). Currently, response to nCRT is most commonly assessed following completion of treatment by physical examination, magnetic resonance imaging or computed tomography scan. Response to treatment is confirmed post-operatively by examination of histologically resected specimens.
There are presently no accurate biomarkers to predict a patient’s response to 5-FU based preoperative neoadjuvant treatment (7). Therefore, patients who have a complete pathologic response with no remaining viable cancer cells may be exposed to unnecessary surgery. Just as importantly, patients who exhibit no pathologic response to 5-FU preoperative treatment are given unnecessary chemotherapy and radiation, with associated toxicities as well as delaying surgery (8).
5-FU metabolism is complex, requiring multiple enzymes, including thymidylate synthase (TS). TS catalyzes the conversion of deoxyuridine monophosphate (dUMP) to thymidine monophosphate (dTMP), which is one of the three nucleotides that forms thymine (9). Thymine is a nucleic acid that is necessary for DNA replication. 5-FU acts to irreversibly inhibit TS, essentially starving rapidly dividing cells by halting DNA production.
Genetic polymorphisms have previously been found in the upstream 5′-promoter region of the TS gene. The two most common TS allelic variations are a double tandem repeat (2R), with a frequency approximately 0.47, and a triple tandem repeat (3R), with a frequency approximately 0.53 (rs34743033) (10). Patients may be homozygous for the 2R allele (2R/2R), homozygous for the 3R allele (3R/3R), or heterozygous (2R/3R) (11). Within the 3R allele, in the second repeat unit, a single nucleotide polymorphism (SNP) may be present from guanine (G) to cysteine (C) (rs2853542) (12). We hypothesize that polymorphisms in the TS gene may affect patients’ pathologic response to preoperative neoadjuvant 5-FU treatment and explain observed clinical differences in response to treatment seen between patients. To our knowledge, this is the first study investigating TS polymorphisms and pathologic response to 5-FU based neoadjuvant therapy in rectal cancer patients.
Methods
Patients
This study was performed on samples from a prospectively-maintained database of a large university colon and rectal surgery practice. The study included consecutive patients from the senior author’s practice who received 5-FU based preoperative nCRT for rectal cancer in whom information was available regarding pre- and post-neoadjuvant staging and who consented for phlebotomy for inclusion in a study focusing on the genetic aspects of gastrointestinal disease. Inclusion criteria were patients with biopsy confirmed rectal adenocarcinoma, who had completed preoperative neoadjuvant 5-FU based chemoradiation and who subsequently underwent radical surgical resection, in whom information was available regarding pre- and post-neoadjuvant staging, and in whom follow-up information was available. Subjects included in this study were recruited from a tertiary referral hospital with a broad geographic referral region. For this reason, preoperative neoadjuvant chemoradiation was given at multiple different sites depending on the patient’s residence and preference. Standard treatment included combination 5-FU and leucovorin chemotherapy with 54 Gy external beam radiation. All pre-neoadjuvant and post-neoadjuvant tumors were staged according to the American Joint Committee on Cancer TNM staging system (13).
Written informed consent was obtained from all patients. The University of Louisville Institutional Review Board approved the study. The patients’ TNM stage, length of time between completion of neoadjuvant chemoradiation and surgery, and time of and status at last follow-up were obtained from office notes, hospital charts, operation notes, endorectal ultrasound (ERUS), colonoscopy, pathology, laboratory and radiology reports (Table 1).
Table 1.
Patient Demographics.
| Patient age | Range | 24 – 88 |
| Mean +/− sd | 58 +/− 14.4 | |
| Median | 57 | |
|
| ||
| AJCC Stage | Pre-neoadjuvant stage | Post-neoadjuvant stage |
|
|
||
| 0 | 0 | 14 |
| I | 4 | 9 |
| II | 17 | 12 |
| A | 15 | 12 |
| B | 2 | 0 |
| C | 0 | 0 |
| III | 23 | 12 |
| A | 5 | 4 |
| B | 18 | 8 |
| C | 0 | 0 |
| IV | 6 | 3 |
| A | 6 | 3 |
| B | 0 | 0 |
|
| ||
| Time from last chemoradiotherapy to surgery (weeks) | Range | 3 – 110 |
| Mean +/− sd | 16 +/− 23.3 | |
| Median | 9 | |
|
| ||
| Length of follow up (months) | Range | 2 – 264 |
| Mean +/− sd | 50 +/− 60.2 | |
| Median | 28 | |
|
| ||
| Status at last follow up | ned* | 25 |
| awd† | 23 | |
| ddd‡ | 1 | |
| du§ | 1 | |
no evidence of disease
alive with disease
died due to disease
died unknown
Each patient’s pre-neoadjuvant cancer stage was compared to his/her post-neoadjuvant cancer stage. Ryan’s three-point grading system (Table 2) was used to quantify patients’ response to nCRT, in order to standardize pathologic response to treatment. Ryan’s three-point scale has a favorable reproducibility and prognostic significance (14).
Table 2.
Ryan Tumor Regression Grade and Patient Pathologic Response.
| Pathologic Response | Ryan Tumor Regression Grade | No. of Patients (%) |
|---|---|---|
| No viable cancer cells or single cells/small groups of cancer cells | 1 | 14 (28) |
| Residual cancer outgrown by fibrosis | 2 | 22 (44) |
| Significant fibrosis outgrown by cancer or no fibrosis with extensive residual cancer | 3 | 14 (28) |
Tumor Regression Grade (TRG) based upon patient pre-operative and post-operative TNM stages using the surgical histopathology. Patients with a score of 1 were considered to have had a complete response (28%), patients with a 2 to have had a partial response (44%), and patients with a score of 3 to have had no response (28%).
DNA Extraction and Whole Genome Amplification
Blood was collected in EDTA-vacutainers (BD, Franklin Lakes, NJ) from patients with rectal cancer who underwent pre-neoadjuvant chemoradiation and stored at 4°C until further use. Genomic DNA was extracted from whole blood samples from using Qiagen® DNeasy Tissue Extraction Kit (Valencia, CA). Whole genome amplification of each sample was performed using the illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Pittsburgh, PA). DNA concentration was determined after whole genome amplification using spectrophotometry.
Genotyping TS
Amplification of the 5′-UTR of the TS gene was performed using the Qiagen® Multiplex Polymerase Chain Reaction (PCR) Kit. PCR of the TS promoter region was carried out with previously described primers (11):
Forward 5′-GTGGCTCCTGCGTTTCCCCC-3′
Reverse 5′-GCTCCGAGCCGGCCACAGGCATGGCGCGG-3′
Each 50 μL PCR reaction contained 1x Qiagen Multiplex PCR Master Mix, 0.2 μL of both the forward and reverse primers, 1x Qiagen Q-Solution, RNase-free water and template DNA. Amplification was performed as follows: 95°C holding stage for 10 min; 37 cycles of denaturation of DNA at 96°C for 1 min, annealing of primers at 60°C for 30 sec, and extension at 72°C for 1 min. The final extension was for 5 min at 72°C incubation. The amplified fragments were separated by size on a 3% agarose gel by electrophoresis, which was stained with Sybr®Gold (Molecular Probes, Eugene, OR).
Restriction Fragment Length Polymorphism (RFLP)
RFLP was performed on samples with at least one 3R allele using RFLP assays as previously described (12). The PCR product from each patient was incubated at 37°C for 18 hours with HaeIII enzyme. The fragment sizes resulting from the RFLP assay were determined using 3% agarose gel electrophoresis stained with Sybr®Gold. A combination of the patient’s original TS genotype and RFLP assay were both used to determine the 3R SNP: 2R/3G, 2R/3C, 3C/3C or 3G/3G.
Results
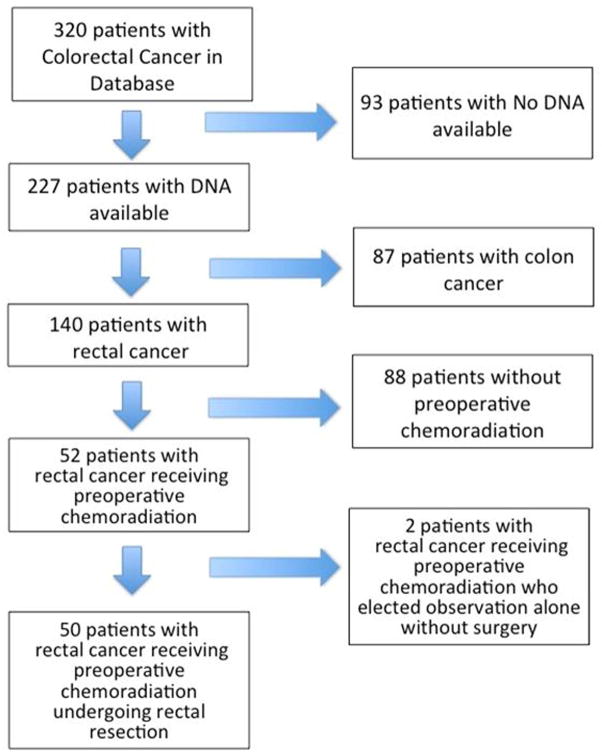
We identified 320 patients from our digestive surgery database with a diagnosis of colorectal cancer. DNA was available for 227 of the 320 patients. Of the patients with available DNA, 140 had a diagnosis of rectal cancer only. Fifty-two of the 140 rectal cancer patients with available DNA met the inclusion criteria and were included in this study. Fifty patients had post-neoadjuvant TNM staging based upon pathology specimens and 2 patients had post-neoadjuvant TNM staging based on clinical staging only (Figure 1).
Figure 1.
Patient Accrual.
Successful DNA extraction, whole genome amplification, and PCR were confirmed by spectrophotometry. Of the 50 patients, 14 (28%) patients were 2R homozygotes, 10 (20%) patients were 3R homozygotes and 26 (52%) patients were heterozygous for 2R/3R.
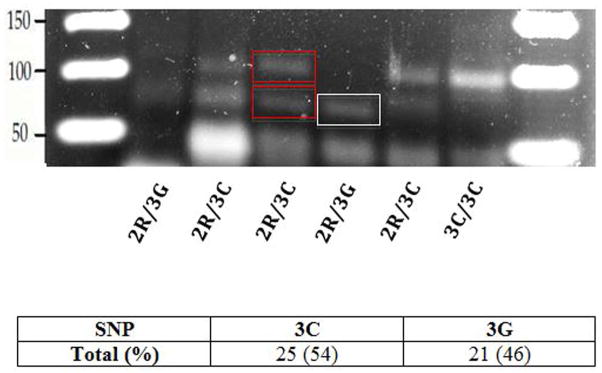
Thirty-six patients with a 3R allele underwent SNP analysis by RFLP. Of the total 46 alleles, SNP analysis identified 54% (25/46) and 46% (21/46) of this population to have the 3C and 3G alleles, respectively (Figure 2). The 3G allele resulted in fragment sizes of 66-bp, while the 3C allele resulted in fragment sizes of both 66- and 94-bp (Figure 2) (12).
Figure 2.
RFLP Gel Electrophoresis. The 3G allele has a digestion band at 66-bp (white box), while the 3C allele has digestion bands at both 94- and 66-bp (red boxes). For each sample, the initial 2R/3R genotype classification and RFLP assay results were compared to determine the TS genotype. RFLP assays revealed that of the 46 3C or 3G alleles studied, 54% were 3C, and 46% were 3G.
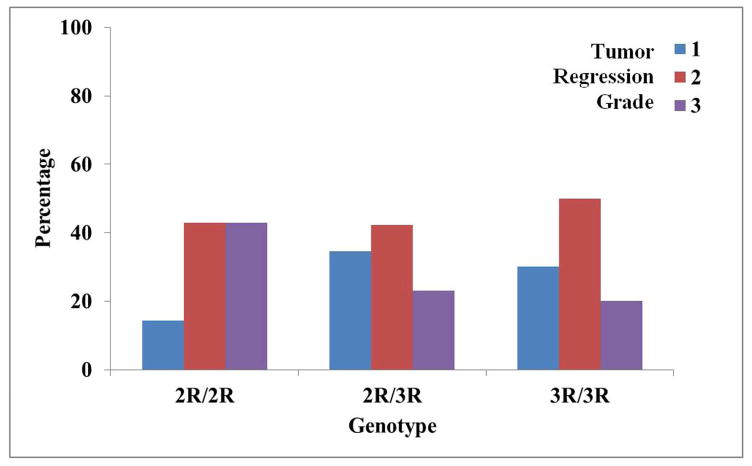
Complete pathologic response occurred in 14 patients (28%), 22 patients (44%) had a partial response and 14 patients (28%) had no response to neoadjuvant chemoradiation, which is consistent with previously published studies (7, 15) with the exception of complete pathologic response that was slightly higher in our study (Table 2). The distribution of TS genotype frequencies as related to tumor regression grade (TRG) is shown in Figure 3.
Figure 3.
TS Genotypes Compared to Tumor Regression Grades (TRG). Each patient received a TRG, from 1–3, based on Ryan’s Tumor Regression Grade (TRG) for his/her pathologic response to preoperative treatment. Of the 14 patients homozygous for the 2R allele, 14% had a TRG of 1, 43% had a TRG of 2 and another 43% had a TRG of 3. Of the 26 heterozygous patients, 35% had a TRG of 1, 42% had a TRG of 2, and 23% had a TRG of 3. Of the 10 patients homozygous for the 3R allele, 30% had a TRG of 1, 50% had a TRG of 2 and 20% had a TRG of 3.
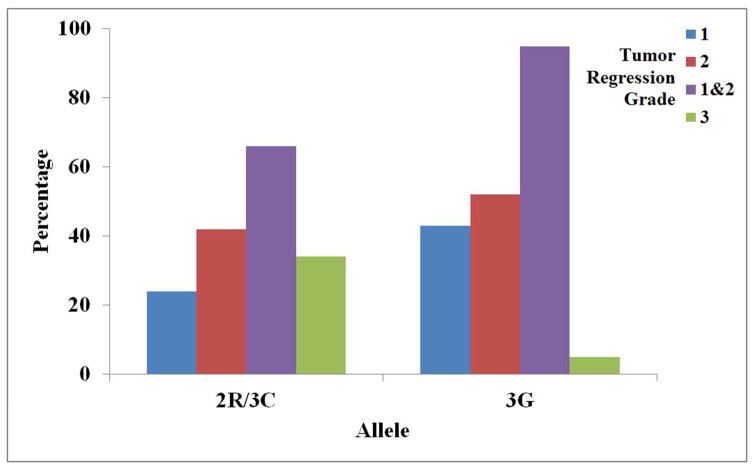
Patients with at least one TS 3G allele were more likely to have complete or partial pathological response to 5-FU neoadjuvant therapy than patients lacking a 3G allele (OR 10.4, 95% CI 1.3–81.6) (p=0.01) (Figure 4 and Table 3). The median time from completion of chemoradiotherapy to surgery was 9 weeks (range 3–110 weeks). The range from nCRT to surgery is exceedingly wide to several outliers. These include a patient with a myocardial infarction who required emergency coronary revascularization and 2 patients who chose to delay surgery significantly following completion of nCRT for personal reasons. There was no significant difference in pathological response to neoadjuvant chemoradiation in patients with at least one TS 3G allele based on time to surgery (<9 weeks OR 7.3, 95% CI 0.9–61.1 p=0.05, >9 weeks OR 7.3, 95% CI 0.4–138.2 p=0.2).
Figure 4.
Allelic Frequencies and Corresponding Tumor Regression Grade. Patients with at least one TS 3G allele were more likely to have complete or partial pathological response to 5-FU neoadjuvant therapy than patients without at least one 3G allele (OR 10.4, 95% CI 1.3–81.6) (p=0.01).
Table 3.
Allelic Frequencies and Corresponding Tumor Regression Grade.
| TS Allele | ||
|---|---|---|
| Tumor Regression Grade | 2R or 3C | 3G |
| 1 | 19 (24) | 9 (43) |
| 2 | 33 (42) | 11 (52) |
| 1 & 2 | 52 (66) | 20 (95) |
| 3 | 27 (34) | 1 (5) |
Tumor Regression Grades can be divided into 1 (complete response), 2 (partial response), 1&2 (any response), and 3 (no response). 20 of the 21 total 3G alleles (95%) were observed in patients with a Tumor Regression Grade of 1 or 2, while only 1 (5%) were observed in patients with a grade of 3. Of the 79 2R and 3C alleles, 66% corresponded to a grade of 1 or 2, while 34% corresponded to a grade of 3.
Many clinical variables, including CEA, were not found to be predictive of response to neoadjuvant chemoradiation. Pre-neoadjuvant CEA levels were available for 41 of 50 patients and were elevated in 7/11 (64%) of responders as compared to 10/30 (33%) of the non-responders (CEA vs. TRG: OR 3.5, 95% CI 0.8–14.8, p=0.15).
Discussion
Preoperative neoadjuvant chemoradiation is the standard treatment for locally advanced rectal adenocarcinoma (16). When compared with post-operative chemoradiation, pre-operative therapy is associated with improved local control, reduced toxicity and increased sphincter preservation (17). Combination 5-FU with leucovorin or oxaliplatin are the chemotherapeutics of choice for preoperative treatment of rectal cancer.
Identification of patients with specific genetic polymorphisms in enzymes involved in 5-FU metabolism, specifically TS, appears to predict the likelihood of pathologic response of rectal cancer to preoperative 5-FU-based neoadjuvant therapy and may alter surgical decision making and aid in selection of preoperative treatment. Another enzyme involved in the metabolism of 5-FU, dihydropyrimidine dehydrogenase (DPD), has also been found to predict patient response to 5-FU treatment (18). DPD is a rate-limiting enzyme in 5-FU metabolism (19), and a study has found patients with decreased enzymatic activity are at a higher risk for developing 5-FU toxicity (20). Approximately 3–5% of the population have a partial DPD deficiency, while 0.2% have a complete deficiency (21). Due to these low frequencies, recruiting a large enough patient sample to develop a screening test for DPD would be difficult. In contrast, the larger frequencies of the different TS genotypes and treatment outcomes makes development of a screening test for TS more feasible. In the future, development of a screening panel for multiple enzymes in the 5-FU metabolic pathway may be an even more accurate prediction of treatment response to neoadjuvant therapy.
One study has shown that a higher number of repeat units in the thymidylate synthase gene leads to greater DNA translation efficiency (22). In vitro studies have shown that the 2R allele produces approximately 3.6 times less mRNA than the 3R allele (23). TS genotypes can be divided into high and low expression types. The 2R/2R, 2R/3C, and 3C/3C genotypes are considered low expression, while the 2R/3G, 3C/3G, and 3G/3G genotypes are considered high expression (12). Each of the high expression genotypes has a 3G allele, meaning that that 3G allele is responsible for a higher expression of TS for 5-FU to target and inhibit. We hypothesized that this particular SNP might be useful in predicting a response in patients with rectal cancer undergoing neoadjuvant therapy since this polymorphism is associated with differences in the amount of functional enzyme produced. The more enzyme present, the more enzyme there is available for 5-FU to act upon, perhaps resulting in improved radiation sensitization.
Patients homozygous for the 3G TS allele (3G/3G) are predicted to have the greatest likelihood of a complete pathological response to 5-FU based nCRT. These patients should respond more favorably to 5-FU than patients with other genotypes. Patients with this genotype may, in theory, even be able to avoid rectal resection. Patients with a complete response that do not undergo surgery should however, continue to have routine monitoring for possible recurrence. Patients with one 3G allele are predicted to have at least a partial pathological response to neoadjuvant chemoradiation. These patients may not be able to avoid rectal resection, however they will benefit from 5-FU based treatment. The presence of the 3G SNP predicts at response (partial or complete) to neoadjuvant therapy and its absence predicts no response. Patients lacking a 3G allele are least likely to respond to 5-FU based nCRT. These patients, specifically patients with at least one 2R allele, are also at a higher risk for early 5-FU toxicity (24). Alternative therapy or bypassing preoperative chemotherapy, which avoids 5-FU toxicity and also prevents delaying necessary surgical intervention, should be considered.
The recommendation for optimal interval between completion of neoadjuvant chemoradiation and surgical intervention has yet to be established (25). Our results show that having at least one 3G allele in the TS gene trends towards a patient’s chance of having at least some pathologic response to neoadjuvant chemoradiation if the surgery is performed within 9 weeks after completing treatment with 5-FU. Increasing the study population may help to better determine the time interval for surgical intervention on completion of nCRT. We suggest that TS genotypes may be used in the future to tailor intervals between neoadjuvant chemoradiation and surgery for rectal cancer.
Conclusions
According to findings of our study, approximately one-third of rectal cancer patients could avoid an unnecessary rectal resection due to complete response to neoadjuvant therapy, and another one-third of patients could bypass potentially harmful preoperative chemotherapy due to lack of response to such therapy as predicted by their TS genotype. It appears that an affordable, efficacious, screening test could be developed based on TS genotyping to predict response to neoadjuvant chemoradiation.
Supplementary Material
Acknowledgments
Funding:
National Cancer Institute grant R25-CA134283 and the John Williamson and Barbara Thruston Atwood Price Family Trust
The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. The authors declare no conflicts of interest.
Footnotes
Author Contributions
Study concept and design: JVC, MRE, SG
Data acquisition: BN, JVC, UN, MRE
Analysis and interpretation of data: BN, JVC, MRE
Drafting of the manuscript: BN, JVC, SG
Critical revision of the manuscript for intellectual content: JVC, UN, MRE, SG
Study supervision: SG
Presented at the 11th Annual Academic Surgical Congress in Jacksonville, FL, February 2–4, 2016
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Casillas S, Pelley RJ, Milson JW. Adjuvant therapy for colorectal cancer: present and future perspectives. Dis Colon Rectum. 1997;40(8):977–92. doi: 10.1007/BF02051209. [DOI] [PubMed] [Google Scholar]
- 3.Maas M, Nelemans PJ, Valentini V, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11(9):835–44. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 4.Weng L, Zhang L, Peng Y, et al. Pharmacogenetics and pharmacogenomics: a bridge to individualized cancer therapy. Pharmacogenomics. 2013;14(3):315–24. doi: 10.2217/pgs.12.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katara P. Single nucleotide polymorphism and its dynamics for pharmacogenomics. Interdiscip Sci. 2014;6(2):85–92. doi: 10.1007/s12539-013-0007-x. [DOI] [PubMed] [Google Scholar]
- 6.McMullen KP, Blackstock AW. Chemoradiation with novel agents for rectal cancer. Clin Colorectal Cancer. 2002;2(1):24–30. doi: 10.3816/CCC.2002.n.008. [DOI] [PubMed] [Google Scholar]
- 7.Ryan J, Warrier S, Lynch A, et al. Predicting pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: A systemic review. Colorectal Disease. 2015 doi: 10.1111/codi.13081. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Papanastasopoulos P, Stebbing J. Molecular Basis for 5-Fluorouracil-related Toxicity: Lessons from Clinical Practice. Anticancer Research. 2014;34(4):1531–5. [PubMed] [Google Scholar]
- 9.Garg D, Henrich S, Salo-Ahen OMH, et al. Novel Approaches for Targeting Thymydiylate Synthase to Overcome the Resistance and Toxicity of Anticancer Drugs. Journal of Medicinal Chemistry. 2010;53:6539–49. doi: 10.1021/jm901869w. [DOI] [PubMed] [Google Scholar]
- 10.Rosmarin D, Palles C, Church D, et al. Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: investigation in the QUASAR2 Study. J Clin Oncol. 2014;32:1031–9. doi: 10.1200/JCO.2013.51.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pullarkat ST, Stoehlamacher J, Ghaderi V, et al. Thymidlyate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. The Pharmacogenomics Journal. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami K, Watanbe G. Identification and Functional Analysis of Single Nucleotide Polymorphism in the Tandem Repeat Seqeunce of Thymidylate Synthase Gene. Cancer Research. 2003;63:6004–7. [PubMed] [Google Scholar]
- 13.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 14.Ryan R, Gibbons D, Hyland JMP, et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology. 2005;47(2):141–6. doi: 10.1111/j.1365-2559.2005.02176.x. [DOI] [PubMed] [Google Scholar]
- 15.Mass M, Nelemans PJ, Valentini V, et al. Long-term outcomes in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2011;11(9):835–44. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Florez LJ, Gomez-Alvarex G, Frunza AM, et al. Predictive markers of response to neoadjuvant therapy in rectal cancer. Journal of Surgical Research. 2014;194(1):120–6. doi: 10.1016/j.jss.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. New England Journal of Medicine. 2004;351(17):1731–40. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 18.Jedrzychowska A, Dolegowska B. Dihydropirymidine dehydrogenase (DPD) -- a toxicity marker for 5-fluorouracil? Ann Acad Med Stetin. 2013;59(2):48–53. [PubMed] [Google Scholar]
- 19.Cai X, Fang JM, Xue P, et al. The role of IVS14+1 G>A genotype detection in the dihydropyrimidine dehydrogenase gene and pharmacokinetic monitoring of 5-fluorouracil in the individualized adjustment of 5-fluorouracil for patients with local advanced and metastatic colorectal cancer. European Review for Medical and Pharmacological Sciences. 2012;18:1247–58. [PubMed] [Google Scholar]
- 20.Milano G, Etienne MC, Pierrefite V, et al. Dihydropyrmidine dehydrogenase deficiency and fluorouracil-related toxicity. Br J Cancer. 1997;79:627–30. doi: 10.1038/sj.bjc.6690098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morel A, Boisdron-Celle M, Fey L, et al. Clinical relevance of different dihydropyrmidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Molecular Cancer Therapeutics. 2006:2895–904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami K, Omura K, Kanehira E, et al. Polymorphic tandem repeats in the thymidylate synthase gene is associated with its protein expression in human gastrointestinal cancers. Anticancer Research. 1999;19:3249–52. [PubMed] [Google Scholar]
- 23.Horie N, Aiba H, Oguro K, et al. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5’-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20:191–7. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 24.Meulendijks D, Jacobs BAW, Aliev A, et al. Increased Risk of Severe Fluoropyrimidine-Associated Toxicity in Patients Carrying a G to C Substitution in the First 28-bp Tandem Repeat of the Thymidylate Synthase 2R Allele. Int J Cancer. 2015;138:245–53. doi: 10.1002/ijc.29694. [DOI] [PubMed] [Google Scholar]
- 25.You KY, Huang R, Zhang LN, et al. Tailored selection of the interval between neoadjuvant chemoradiotherapy and surgery for locally advanced rectal cancer: analysis based on the pathological stage or chemoradiation response. J Cancer Res Clin Oncol. 2015;141(4):719–28. doi: 10.1007/s00432-014-1843-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.