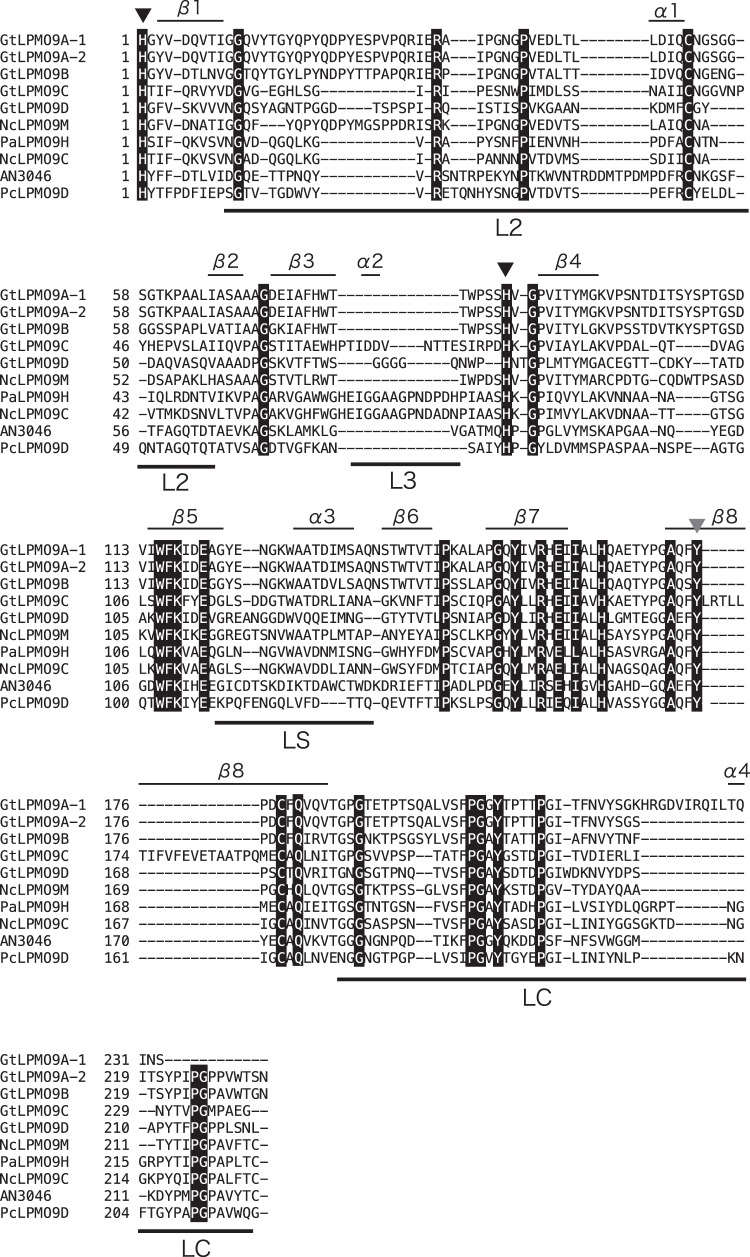
FIG 2.
Multiple-sequence alignment of the catalytic domains of selected LPMO9s. Fully conserved residues are printed in white on a black background. Active-site histidines (black-filled triangles) and a tyrosine (gray-filled triangle) involved in copper coordination are indicated. The labeled bars over the sequences indicate known variable regions in LPMO9s (15, 51). Note that GtLPMO9A-2 and GtLPMO9D have C-terminal extensions; for more information, see Fig. S2 to S4 in the supplemental material. This alignment was generated using MAFFT, version 7.295 (39), available at the European Bioinformatics Institute website (https://www.ebi.ac.uk/Tools/msa/mafft/).