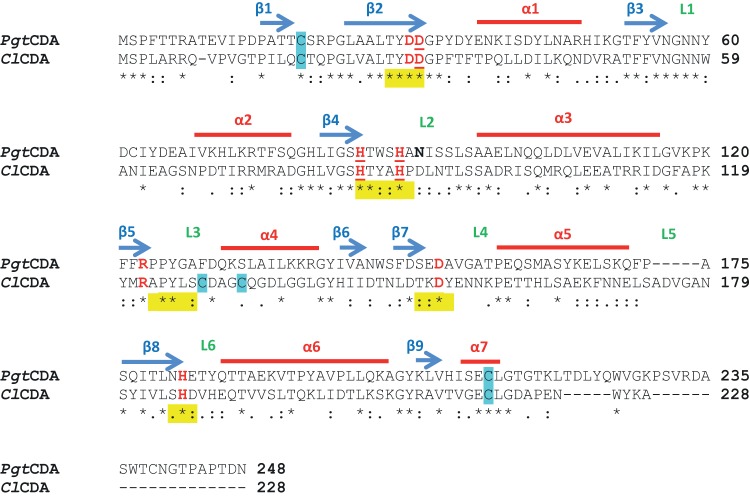
FIG 1.
Clustal Omega amino acid sequence alignment of PgtCDA from P. graminis and ClCDA (PDB 2IWO) from C. lindemuthianum. The sequence analysis shows comparison of amino acid sequences of ClCDA and PgtCDA without signal peptide removed using SignalP 3.0 server. Alpha-helices and beta-sheets are marked by red lines and blue arrows, respectively, and the positions of loops (L) decorating the active site are indicated in green. The five conserved motifs of CE4 enzymes are marked in yellow. The catalytic and metal ligand binding (underlined) residues are marked in red. The cysteines predicted in disulfide bonds are marked in blue. One potential N-glycosylation site is marked in bold. The consensus symbols have the usual meanings, i.e., asterisks (*) indicate positions which have a single, fully conserved residue, colons (:) indicate conservation between groups of strongly similar properties, and periods (.) indicate conservation between groups of weakly similar properties.